Abstract
Comment on: Tan AY, et al. Proc Natl Acad Sci USA 2012; 109:6030-5.
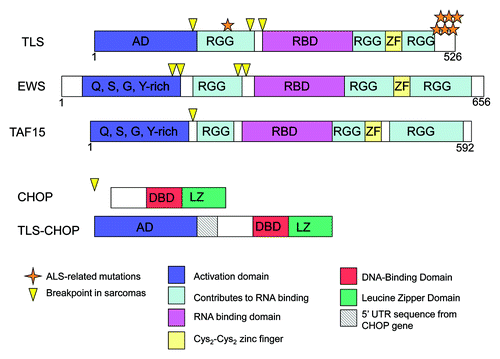
Translocated in liposarcoma/fused in sarcoma (TLS/FUS, or TLS) was discovered 20 y ago through a characteristic chromosomal translocation.Citation1 TLS is related to EWS (Ewing sarcoma) and TAF15 (TBP-associated factor 15). All are abundant nuclear proteins with an N-terminal transcriptional activation domain, an RNA-binding domain, RGG boxes and a zinc finger (). The N terminus of each has been found fused to a transcription factor, e.g., CHOP, through chromosomal translocation in a variety of sarcomas (), resulting in deregulated expression of the transcription factor’s target genes. While the fusion proteins and their cancer phenotypes had been studied, the role of native TLS is not well understood, although a number of roles have been suggested, including functions in transcription and mRNA splicing. Furthermore, interest in these proteins has increased considerably with the discovery that TLS is a frequent target of mutation in both sporadic and familial forms of amyotrophic lateral sclerosis (ALS, or Lou Gehrig disease).Citation2,Citation3
Figure 1. Structures of TLS, EWS, TAF15, CHOP and TLS-CHOP. Domains of TLS, CHOP and TLS-CHOP are depicted, including the activation domain (AD), RGG boxes that contribute to RNA binding (RGG), RNA-binding domain (RBD), Cys2-Cys2 zinc finger (ZF), DNA-binding domain (DBD) and leucine zipper domain (LZ). The location of sarcoma breakpoints (arrowheads) and ALS-related missense mutations in TLS (stars) are also depicted.
Seemingly simple questions are the identity of TLS target genes, and how they are recognized. TLS has previously been shown to be able to bind both DNA and RNA, but whether it uses this ability, as opposed to protein-protein interactions, to associate with target genes, is not known.Citation1 To address this question, we performed chromatin immunoprecipitation (ChIP) with HeLa cells and a human promoter microarray to identify promoters that are bound, directly or indirectly, by TLS.Citation4 We found a number of genes, ~50 with p value < 0.01, over 1,000 with p value < 0.05, associated with TLS. We verified several, and showed changes in expression, both positive and negative, when TLS levels were altered. A bioinformatics analysis of the microarray data identified three statistically significant sequences enriched in DNA bound by TLS, and one or more of these sequences were present in the promoter regions of the genes we verified. Surprisingly, we found that purified TLS bound these sequences with specificity as single-strand, but not double-strand, DNA. What domain(s) of the protein is (are) involved in DNA binding, and how this relates to TLS promoter recognition in vivo, are important goals for the future.
Several of the genes we identified are involved in neuronal functions. Attention to the role of TLS in neurons has intensified since the discovery that TLS is an important ALS target gene. Most mutations are in the C terminal domain () and result in altered localization, such that all or a significant fraction of the protein accumulates in the cytoplasm, largely in aggregates. Arginine methylation at R521, a frequent site of mutation, or other arginine residues, may affect cellular localization and/or protein-protein interactions. Another nuclear protein, TDP-43 (TAR DNA-binding protein 43), which has similar domains to TLS, was also found to be a target of mutations in ALS.Citation5 In this case, mutant proteins are modified by hyperphosphorylation and ubiquitination and also aggregate in the cytoplasm. Thus, incorrect localization or altered post-translational modification and aggregation may result in the motor neuron degeneration that occurs in ALS. A critical question is how these aggregates lead to disease. One possibility is that the decrease in nuclear TLS is important, altering the expression of target genes. If so, it will be important to determine if genes we identified are misexpressed in ALS.
One of the TLS target genes we found is ZNF294, or LISTERIN, which encodes an E3 ubiquitin ligase mutated in some colon cancer cell lines,Citation6 and which is downregulated by TLS. The yeast homolog, Ltn1, marks nascent proteins translated from mRNA lacking a stop codon for degradation, while mutation of the murine homolog causes neurodegeneration in mice.Citation7,Citation8 While the causative genes for many neurological diseases have not been mapped, the involvement of LISTERIN in neurodegeneration, and its regulation by TLS, indicates that this pathway may be involved in ALS. Thus, TLS regulates a gene involved in control of protein quality, and disruption of this pathway may contribute to neurodegenerative disease.
MECP2 (encoding methyl CpG-binding protein 2) is another TLS target gene, in this case activated by TLS, with a role in neurological disorders. MECP2 was originally thought to act as a transcriptional repressor but was also found to activate a number of genes.Citation9 Mutations cause the neurodevelopmental disorder Rett syndrome, while MECP2 overexpression results in other neurological ailments.Citation10 Intriguingly, TDP-43 binds MECP2 protein, while TLS acts at the transcriptional level in regulating MECP2. Thus, TLS and TDP-43 may regulate a common pathway important in neurological disorders.
Increasing our understanding of how TLS, as well as EWS and TAF15 (the “TET proteins”), regulates gene expression, will have implications for both cancer and neurodegenerative diseases. With respect to cancer, it seems clear that creation of oncogenic fusion transcription factors is the major culprit, but it is conceivable that changes in expression of natural TET target genes, reflecting decreased levels of the native protein, may play some role, and it will be of interest to determine whether expression of TLS target genes varies in TLS-derived sarcomas. Likewise, does expression of TLS target genes vary in ALS, perhaps reflecting decreased levels of the protein in neuronal cell nuclei? These and other questions will likely maintain interest in TET proteins for some time.
References
- Tan AY, et al. J Mol Cell Biol 2009; 1:82 - 92; http://dx.doi.org/10.1093/jmcb/mjp025; PMID: 19783543
- Strong MJ, et al. FEBS J 2011; 278:3569 - 77; http://dx.doi.org/10.1111/j.1742-4658.2011.08277.x; PMID: 21810174
- Lagier-Tourenne C, et al. Cell 2009; 136:1001 - 4; http://dx.doi.org/10.1016/j.cell.2009.03.006; PMID: 19303844
- Tan AY, et al. Proc Natl Acad Sci USA 2012; 109:6030 - 5; http://dx.doi.org/10.1073/pnas.1203028109; PMID: 22460799
- Neumann M, et al. Science 2006; 314:130 - 3; http://dx.doi.org/10.1126/science.1134108; PMID: 17023659
- Ivanov I, et al. Oncogene 2007; 26:2873 - 84; http://dx.doi.org/10.1038/sj.onc.1210098; PMID: 17086209
- Bengtson MH, et al. Nature 2010; 467:470 - 3; http://dx.doi.org/10.1038/nature09371; PMID: 20835226
- Chu J, et al. Proc Natl Acad Sci USA 2009; 106:2097 - 103; http://dx.doi.org/10.1073/pnas.0812819106; PMID: 19196968
- Chahrour M, et al. Science 2008; 320:1224 - 9; http://dx.doi.org/10.1126/science.1153252; PMID: 18511691
- Zachariah RM, et al. Neural Plast 2012; PMID: 22474603
- Sephton CF, et al. J Biol Chem 2011; 286:1204 - 15; http://dx.doi.org/10.1074/jbc.M110.190884; PMID: 21051541