Abstract
Histone modifications and their modifying enzymes are fundamentally involved in the epigenetic regulation of adipogenesis. This study aimed to define the roles of various histone modifications and their “division of labor” in fat cell differentiation. To achieve these goals, we examined the distribution patterns of eight core histone modifications at five key adipogenic regulatory genes, Pref-1, C/EBPβ, C/EBPα, PPARγ2 and aP2, during the adipogenesis of C3H 10T1/2 mouse mesenchymal stem cells (MSCs) and 3T3-L1 preadipocytes. We found that the examined histone modifications are globally stable throughout adipogenesis but show distinct and highly dynamic distribution patterns at specific genes. For example, the Pref-1 gene has lower levels of active chromatin markers and significantly higher H3 K27 tri-methylation in MSCs compared with committed preadipocytes; the C/EBPβ gene is enriched in active chromatin markers at its 3′-UTR; the C/EBPα gene is predominantly marked by H3 K27 tri-methylation in adipogenic precursor cells, and this repressive marker decreases dramatically upon induction; the PPARγ2 and aP2 genes show increased histone acetylation on both H3 and H4 tails during adipogenesis. Further functional studies revealed that the decreased level of H3 K27 tri-methylation leads to de-repression of Pref-1 gene, while the increased level of histone acetylation activates the transcription of PPARγ2 and aP2 genes. Moreover, the active histone modification-marked 3′-UTR of C/EBPβ gene was demonstrated as a strong enhancer element by luciferase assay. Our results indicate that histone modifications are gene-specific at adipogenic regulator genes, and they play distinct roles in regulating the transcriptional network during adipogenesis.
Introduction
Mammalian cells store genetic information within chromatin and control gene expression by modulating chromatin structure. Chromatin is mainly composed of double-stranded DNA wrapped around core histones, which are targets of various posttranslational modifications, including acetylation, methylation and phosphorylation.Citation1 These histone modifications are fundamentally involved in gene regulation by either directly modulating chromatin structureCitation2,Citation3 or providing binding platforms for activators or repressors of gene transcription.Citation4,Citation5 Among the different forms of histone modification, acetylation is generally associated with active gene transcription,Citation3,Citation6 while the role of methylation depends specifically on the siteCitation7,Citation8 and degree of methylation (i.e., mono-, di- or tri-methylation).Citation9,Citation10 For instance, mono-methylation of histone H3 lysine (K) 4 has been shown to mark enhancer elements in the human genome, and tri-methylation is mainly found at active promoters.Citation11 In contrast, H3 K9 and K27 tri-methylation have been defined as repressive markers. K9 tri-methylation recruits heterochromatin-associated protein-1 (HP1) to establish heterochromatic regions,Citation5 while K27 tri-methylation is involved in PcG-mediated gene silencing.Citation12 Although each individual histone modification has its own function, different modifications also work in combination to fine-tune gene expression. It has been proposed that in human embryonic stem (ES) cells, an active marker (H3 K4 tri-methylation) and a repressive marker (H3 K27 tri-methylation) define a bivalent chromatin domain that silences key developmental genes while keeping them poised for activation upon differentiation.Citation13 Corresponding to the large number of histone modifications, there are numerous histone-modifying enzymes with distinct histone substrate preferences and biological functions.Citation14 For example, acetylation of the histone H3 N-terminal tail is mainly catalyzed by the lysine acetyltransferase (KAT) Gcn5/PCAF (KAT2A/2B) as well as p300/CBP (KAT3A/3B), and the H4 tail is predominantly acetylated by the MYST family of KATs.Citation15 The opposing activity, removal of histone acetylation, is performed by histone deacetylase (HDAC) 1–11 and sirtuin 1–7 in mammals. In contrast to the KATs, which usually target multiple sites, lysine methyltransferases (KMT) have restricted substrate preferences. The MLL family of KMTs methylates histone H3 K4, and Ezh2 is responsible for adding methyl groups to H3 K27. Histone methylation was once thought to be irreversible, but since the discovery of the first lysine demethylase (KDM), LSD1 (KDM1),Citation16 a large group of JmjC domain-containing KDMs have been identified.Citation17 As a major mechanism of epigenetic gene regulation, histone modifications have been extensively studied in the last decade, and roles have been identified for these modifications in diverse cellular processes, including mitosis, programmed cell death and differentiation. One such process of special interest to our group is adipogenesis (fat cell differentiation), which is directly linked to human obesity and related diseases such as diabetes.
Adipogenesis is a complex physiological process that requires the concerted regulation of gene expression by various adipogenic factors. Among these factors, PPAR-γ and C/EBPα play central roles by controlling a large number of downstream adipogenic genes during the differentiation process.Citation18 Of these, PPAR-γ, a member of the hormone nuclear receptor super-family, is the only factor that is both necessary and sufficient to promote adipogenesis.Citation19,Citation20 Mature adipocytes are differentiated from preadipocytes, which are originally derived from multipotent mesenchymal stem cells (MSCs). When given the appropriate environmental cues, MSCs give rise to multiple lineages, and Wnt signaling plays a negative role in adipocyte lineage commitment.Citation21 Another repressor of adipogenesis is preadipocyte factor-1 (Pref-1, also called Dlk1), an epidermal growth factor-repeat containing transmembrane protein that is specifically expressed in preadipocytes but not in mature fat cells.Citation22 Pref-1 represses adipogenesis by upregulating Sox9 expression, which subsequently suppresses the expression of the early adipogenic genes C/EBPβ and C/EBPδ.Citation23 The expression of C/EBPβ and C/EBPδ is quickly induced after treatment with dexamethasone (DEX) and 3-isobutyl-1-methylxanthine (IBMX), with their gene products subsequently cooperating to activate the expression of PPAR-γ2 and C/EBPα, which establishes terminal adipogenesis.Citation24,Citation25 The induced expression of C/EBPβ and C/EBPδ precedes PPAR-γ2 and C/EBPα expression by 1–2 d, suggesting that additional steps are required to fully activate PPAR-γ2 and C/EBPα transcription. One explanation for this lag is that binding of C/EBPβ to the C/EBPα promoter requires changes in chromatin structure that occur during clonal expansion and terminal differentiation.Citation26 After induction, PPAR-γ and C/EBPα form a positive feedback loop, each facilitating the expression of the other. This feedback loop leads to the activation of the entire adipogenic program and the expression of a wide range of factors required for mature adipocyte function. The adipose-specific fatty acid binding protein aP2/FABP4 is one such factor and plays an essential role in cellular fatty acid binding and transportation.
Histone modifications and their corresponding enzymes are clearly involved in both the determination and the differentiation processes of adipogenesis. For instance, histone H3 K4 di-methylation has been shown to be a marker of transcriptional competence for the adiponectin gene in preadipocytes.Citation27 Recently, the same group showed that the histone H3 K4/K9 demethylase LSD1 promotes adipogenesis by modulating H3 K4/K9 methylation at the C/EBPα promoter.Citation28 In addition, certain histone methylases promote adipogenesis, MLL3 through H3 K4 tri-methylationCitation29 and PR-Set7/Set8 through H4 K20 mono-methylation.Citation30 A recent report further showed that Ezh2 represses Wnt signaling through H3 K27 tri-methylation to facilitate adipogenesis.Citation31 Moreover, the histone H3 K9 demethylase Jhdm2a was previously shown to have a role in obesity resistance.Citation32 In addition to directly binding adipogenic regulator genes, histone-modifying enzymes also modulate adipogenesis by interacting with adipogenic regulators. One example is the representative member of the mammalian sirtuin family, SIRT1, which attenuates adipogenesis by binding to the master regulator PPAR-γ and repressing its target genes upon food limitation.Citation33 On the other hand, the KATs p300/CBPCitation34 and Tip60Citation35 promote adipogenic gene activation through direct interaction with PPAR-γ, enhancing its transcriptional activity.Citation35
To further elucidate the essential role of histone modification in the epigenetic regulation of adipogenesis, we systematically examined the distribution patterns of eight histone modifications using chromatin immunoprecipitation (ChIP) at five key adipogenic regulatory genes, Pref-1, C/EBPβ, C/EBPα, PPARγ2 and aP2, during the adipogenesis of C3H 10T1/2 mouse MSCs and 3T3-L1 preadipocytes. These genes were chosen because of their significant roles in adipogenesis and also because they represent examples of repressed (Pref-1), early activated (C/EBPβ) and middle-late-induced adipogenic regulator genes (C/EBPα, PPARγ2 and aP2). The histone modifications examined include H3 K4 mono-, di- and tri-methylation, H3 K9 tri-methylation, H3 K27 tri-methylation, H4 K20 mono-methylation, H3 K9/K14 acetylation and H4 K12 acetylation. Here, we show that the patterns of these histone modifications are distinct at different genes and highly dynamic during adipogenesis. Modulating these gene-specific histone modifications has a direct impact on corresponding gene transcription. By analyzing these histone modification patterns, a strong enhancer element was identified for C/EBPβ gene. Our results demonstrate that histone modifications and modifying enzymes have a highly specific “division of labor” in regulating various adipogenic regulatory genes during fat cell differentiation.
Results
Expression levels of key adipogenic regulatory genes except for Pref-1 are largely identical in 3T3-L1 and 10T1/2 cells
The 3T3-L1 and 10T1/2 murine cell lines have been well-established as in vitro models of adipogenesis. Using RT-qPCR, we examined the gene expression profiles of several key adipogenic regulatory genes, Pref-1, C/EBPβ, C/EBPα, PPARγ2 and aP2, in both cell lines at different stages of adipogenesis. Consistent with previous reports, the patterns of gene expression for C/EBPβ, C/EBPα, PPARγ2 and aP2 in both cell lines are almost identical (Fig. S1B–E). For Pref-1, the mRNA level is significantly higher in 3T3-L1 preadipocytes compared with 10T1/2 MSCs at the beginning of adipogenesis (Fig. S1A).Citation36 Pref-1 expression then decreases more than 90% in the 48 h following induction. Interestingly, although the starting levels of Pref-1 transcription differ between 3T3-L1 and 10T1/2 cells, their decreasing trends are similar (Fig. S2). C/EBPβ expression quickly follows adipogenic signals, reaching its peak several hours after induction. After a lag of about 2 d, the C/EBPα, PPARγ2 and aP2 genes are activated to fully establish terminal adipogenesis.
Global levels of histone modifications remain stable during adipogenesis
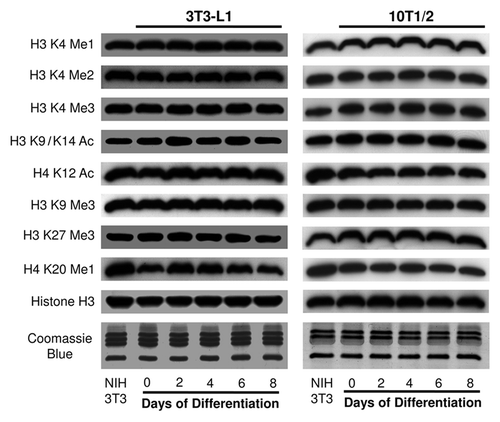
Given that histone modifications are fundamentally involved in adipogenesis, we first examined the state of eight core histone modifications at the global level using western blotting (). Core histones were isolated from 3T3-L1 and 10T1/2 cells at different time points during adipogenesis, and specific antibodies against histone H3 K4 mono-, di- and tri-methylation, H3 K9/K14 acetylation, H4 K12 acetylation, H3 K9 tri-methylation, H3 K27 tri-methylation and H4 K20 mono-methylation were used to evaluate the bulk levels of the corresponding histone modifications. To compare the histone modification states of 3T3-L1 and 10T1/2 cells, we also purified core histones from NIH 3T3 cells and used this sample as an internal control. As shown in , most of the examined histone modifications are stable at bulk levels during adipogenesis in both cell lines. The chromatin modification states at these sites are comparable between 3T3-L1 and 10T1/2 cells when normalized to the NIH 3T3 results. We observed a modest increase in H4 K20 mono-methylation from day 0 to day 2 of differentiation in 3T3-L1 cells, but this pattern was not observed in 10T1/2 cells. Coomassie blue-stained gels and histone H3 western blotting were used to show equal loading in this assay. These results demonstrate that the examined histone modifications are not affected at the global level by adipogenic differentiation.
Figure 1. Global levels of histone modifications remain stable during adipogenesis. Global levels of histone modifications were determined by western blotting in both 3T3-L1 and 10T1/2 cells. Histone samples were collected at the indicated time points during adipogenesis. Samples from NIH 3T3 cells were used as an internal control for comparisons between the 3T3-L1 and 10T1/2 samples. The examined histone methylations were H3 K4 Me1, Me2 and Me3; K9 Me3; K27 Me3 and H4 K20 Me1. The examined histone acetylations were H3 K9/K14 Ac and H4 K12 Ac. Histone H3 western blots and Coomassie blue-stained gels demonstrated equal loading.
General histone loss occurs in the clonal expansion stage of adipogenesis
Histones have been demonstrated to be gene regulators,Citation37 and active gene transcription is usually accompanied by histone loss at the promoter region.Citation38,Citation39 We examined the distribution pattern of histone H3 at specific adipogenic regulatory genes, Pref-1, C/EBPβ, C/EBPα, PPARγ2 and aP2, using ChIP assays. For each gene, we selected four regions to examine. They are regions 2 kb and 1 kb upstream of TSS as well as promoter and exon. For C/EBPα and C/EBPβ genes, we also examined their conserved 3′-UTRs identified through sequence conservation analysis (Fig. S3). In addition, we included representative silent (Chr. 15) and active (GAPDH) chromatin regions as controls in the ChIP-qPCR experiments (Experimental Procedures). To our surprise, we did not observe significant histone loss during the activation of adipogenic genes. Instead, we found a general loss of histones at all of the examined regions at the hour 6 and day 1 time points (Figs. S4B and 5B). This time frame coincides with the clonal expansion stage of adipogenesis. Adipogenic precursor cells must go through the clonal expansion stage after contact growth inhibition to fully establish terminal adipogenesis. Usually this stage starts on day 0 and ends on day 2 after induction. During this stage, cells reenter the cell cycle from growth arrest and undergo 2–3 rounds of mitosis. We interpret the general histone loss at hour 6 and day 1 as the result of cell cycle-related fluctuation in histone levels in the chromatin fraction. We also found that the GAPDH promoter has less histone H3 occupancy than the silent region on Chr.15, which correlates negatively with gene activity. Negative control experiments were performed in parallel using rabbit IgG, and we observed baseline signals for all of the examined regions in these assays (Figs. S4A and 5A).
Patterns of histone H3 K4 methylation at adipogenic regulatory genes
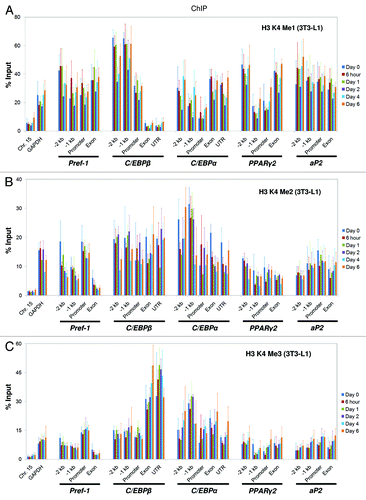
Histone H3 K4 can be mono-, di- or tri-methylated, and each form of methylation has specific biological functions. To explore the roles of H3 K4 methylation in adipogenesis, we examined distribution patterns for the three types of methylation using ChIP assays. We found that the C/EBPβ exon and 3′-UTR lack significant amounts of K4 mono-methylation and, interestingly, are highly enriched for K4 tri-methylation (; Fig. S6A and C). As the three forms of methylation are mutually exclusive at the same site, the basal level of K4 mono-methylation may allow the tri-methylated form to build up at specific genomic loci to facilitate gene activation. We also detected increasing levels of K4 tri-methylation at all of the induced adipogenic genes (; Fig. S6C), which positively correlate with gene activity. The most obvious increasing trend was observed at the aP2 gene in 10T1/2 cells (Fig. S6C). The patterns of K4 methylation in 10T1/2 cells largely resemble the patterns in 3T3-L1 cells, except at the Pref-1 gene, where the levels of all three forms of methylation are lower in 10T1/2 cells (~50% of 3T3-L1 signal). This difference in methylation coincides with the difference in gene expression. In control experiments, we found that all three forms of K4 methylation are much more abundant at the GAPDH promoter compared with the Chr.15 region (; Fig. S6). The most significant difference occurs with K4 di-methylation, for which the relative enrichment at GAPDH is nearly 10-fold that of Chr.15 (; Fig. S6B). We conclude that H3 K4 tri-methylation positively correlates with gene activation and that it marks the C/EBPβ exon and 3′-UTR during adipogenesis.
Figure 2. Histone H3 K4 methylation states at the genomic loci of key adipogenic regulators during 3T3-L1 adipogenesis. Levels of histone H3 K4 (A) mono-methylation, (B) di-methylation and (C) tri-methylation at the genomic loci of the key adipogenic regulators Pref-1, C/EBPβ, C/EBPα, PPARγ2 and aP2 during the adipogenesis of 3T3-L1 cells were examined by ChIP analysis using specific antibodies. The ChIP-qPCR primers used in this study were as follows: Chr. 15, which targets a silent region on mouse chromosome 15 containing no known gene within 500 kb; GAPDH, which targets the promoter region of the actively transcribed GAPDH gene and -2 kb and -1 kb primers, which target regions 2 kb and 1 kb upstream of the TSS, respectively. Promoter primers target regions within 500 bp upstream of the TSS. UTR primers were designed to target large conserved 3′-UTRs in the C/EBPα and C/EBPβ gene sequences. Details of these primers are given in the Experimental Procedures section. ChIP samples were collected at the indicated time points during adipogenesis. These results are the averages of three independent ChIP-qPCR assays, and the error bars indicate standard deviations.
Distinct patterns of histone acetylation on H3 and H4 tails
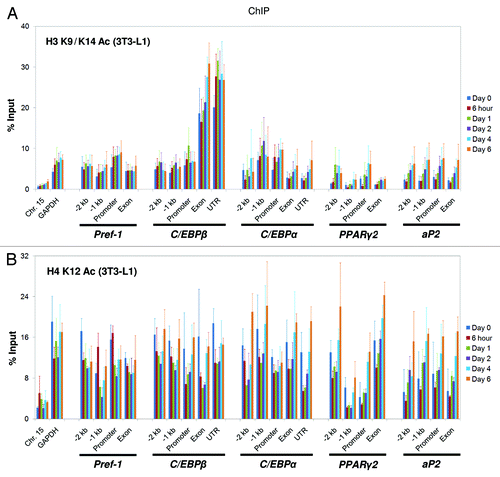
Histone H3 and H4 tails are heavily acetylated at multiple sites. These acetylation sites are generally linked to gene activation, and their functions were assumed to be largely redundant. In this study, we aimed to distinguish the roles of these two histone tails in adipogenesis, and we selected H3 K9/K14 acetylation and H4 K12 acetylation to represent H3 and H4 acetylation, respectively. We used ChIP assays to examine the acetylation of H3 and H4 at the five adipogenic regulatory genes during differentiation. To our surprise, we found that the distribution patterns of H3 and H4 acetylation are distinct, suggesting that these two histone tails play different roles in regulating adipogenic gene expression. Like H3 K4 tri-methylation, H3 K9/K14 acetylation showed the highest enrichment at the C/EBPβ exon and 3′-UTR (; Fig. S7A), but this pattern was not observed for H4 K12 acetylation (; Fig. S7B). Histone acetylation is well-established as a marker of active gene transcription, and we confirmed this relationship by showing that both H3 and H4 acetylation correlate positively with the induction of adipogenic genes (Figs. S1, 3 and 7). Among all of the examined genes, the increase in histone acetylation levels is most evident at the aP2 gene (; Fig. S7). The patterns of both H3 K9/K14 acetylation and H4 K12 acetylation in 10T1/2 cells are similar to the patterns in 3T3-L1 cells, except for the Pref-1 gene, where both the H3 and the H4 acetylation levels were about half of that in 3T3-L1 cells. At the control regions, we observed 4~5-fold higher levels of both H3 and H4 acetylation at the active GAPDH promoter compared with the silent Chr.15 region (; Fig. S7).
Figure 3. Distinct patterns of histone acetylation on the H3 and H4 tails at the genomic loci of key adipogenic regulators during 3T3-L1 adipogenesis. Levels of histone (A) H3 K9/K14 acetylation and (B) H4 K12 acetylation at the genomic loci of the key adipogenic regulators Pref-1, C/EBPβ, C/EBPα, PPARγ2 and aP2 during the adipogenesis of 3T3-L1 cells were examined by ChIP analysis using specific antibodies. The primers used in this study are described in . ChIP samples were collected at the indicated time points. These results are the averages of three independent ChIP-qPCRs, and the error bars indicate standard deviations.
Dynamic pattern of the histone-repressive marker H3 K27 tri-methylation during adipogenesis
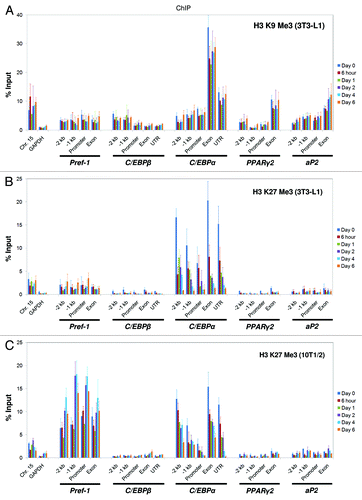
Depending on the site and degree, histone methylation can lead to gene activation or repression. Tri-methylated histone H3 K9 and tri-methylated H3 K27 are well-characterized as repressive markers, although they exert their functions through different mechanisms. We examined the distribution patterns of these two modifications using ChIP analysis during adipogenesis. In 3T3-L1 cells, we found that both K9 and K27 tri-methylation mark the C/EBPα gene (), but K9 tri-methylation is enriched at the exon and 3′-UTR and is relatively stable during C/EBPα gene activation. In contrast, K27 tri-methylation marks the entire C/EBPα gene before induction and decreases dramatically as the gene is activated during adipogenesis (Figs. S1 and 4B). In 10T1/2 cells, the pattern of K9 tri-methylation is almost identical to that of 3T3-L1 cells (Fig. S8A), but we observed a significant difference for K27 tri-methylation at the Pref-1 gene (compare ). At the Pref-1 gene, the level of K27 tri-methylation is 5~10-fold higher in 10T1/2 cells than in 3T3-L1 cells, and its dynamic change negatively correlates with gene activity (Figs. S2B and 4C). At the C/EBPα gene, K27 tri-methylation shows the same decreasing pattern in 10T1/2 cells as in 3T3-L1 cells. Consistent with their roles as repressive markers, H3 K9 and K27 tri-methylation are enriched at the silent region in Chr.15 but not at the active GAPDH gene (; Fig. S8A).
Figure 4. Histone H3 K9 and K27 tri-methylation states at the genomic loci of key adipogenic regulators during 3T3-L1 and C3H 10T1/2 adipogenesis. Levels of histone (A) H3 K9 tri-methylation and (B) H3 K27 tri-methylation were examined by ChIP analysis at the genomic loci of the key adipogenic regulators Pref-1, C/EBPβ, C/EBPα, PPARγ2 and aP2 during the adipogenesis of 3T3-L1 cells. (C) The H3 K27 tri-methylation state during the adipogenesis of C3H 10T1/2 cells. The primers used in this study are described in . ChIP samples were collected at the indicated time points. These results are the averages of three independent ChIP-qPCRs, and the error bars indicate standard deviations.
Histone H4 K20 mono-methylation marks the C/EBPβ gene
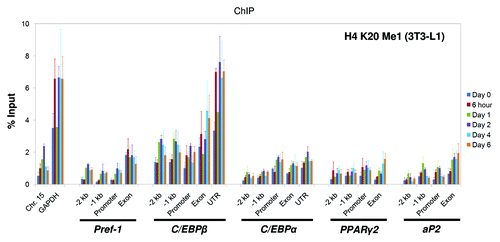
Histone H4 K20 mono-methylation was recently shown to be involved in regulating adipogenesis.Citation30 We determined its occupancy at the adipogenic regulatory genes using ChIP assays. We found that H4 K20 mono-methylation selectively marks the C/EBPβ exon and 3′-UTR, similar to H3 K4 tri-methylation and K9/K14 acetylation (; Fig. S8B). Regarding the control regions, the active GAPDH promoter has higher levels of this modification when compared with the silent region on Chr.15 (; Fig. S8B).
Figure 5. Histone H4 K20 mono-methylation states at the genomic loci of key adipogenic regulators during 3T3-L1 adipogenesis. Levels of histone H4 K20 mono-methylation at the genomic loci of the key adipogenic regulators Pref-1, C/EBPβ, C/EBPα, PPARγ2 and aP2 during the adipogenesis of 3T3-L1 cells were examined by ChIP analysis as described above. The primers used in this study are described in . ChIP samples were collected at the indicated time points. These results are the averages of three independent ChIP-qPCRs, and the error bars indicate standard deviations.
Modulating specific histone modifications has a direct impact on the transcriptional activity of adipogenesis regulatory genes
After obtaining the distribution patterns of various histone modifications at Pref-1, C/EBPβ, C/EBPα, PPARγ2 and aP2 genes, we next asked whether these patterns can guide us to identify the specific epigenetic mechanism that controls the expression of individual adipogenesis regulatory gene. Given that the expression of most adipogenic genes are interconnected and dependent on the progress of adipogenesis, we chose to examine the direct impact of histone modification change on the respective gene activity in preadipocytes, where the adipogenic program hasn't been established, to avoid the complexity of indirect effects.
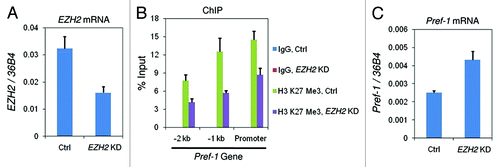
At the Pref-1 gene, we had observed a predominant enrichment of H3 K27 tri-methylation in 10T1/2 MSCs compared with the committed 3T3-L1 preadipocytes (compare ). This is anti-correlated with the gene expression profile of Pref-1 (Fig. S1A), suggesting that this repressive marker is involved in silencing the Pref-1 gene in 10T1/2 cells. To test this hypothesis, we knocked down the K27 methylase Ezh2 in 10T1/2 cells using siRNA. As shown in , EZH2 mRNA was reduced to about 50% of the amount in control cells. Using ChIP assay, we found that the level of K27 tri-methylation at the Pref-1 promoter decreased 40–60% after EZH2 siRNA treatment (). And this is accompanied by a 2-fold increase of Pref-1 gene expression ().
Figure 6. The decreased level of H3 K27 tri-methylation leads to de-repression of Pref-1 gene. Non-targeting control siRNA (Ctrl) or EZH2-targeting siRNA (EZH2 KD) were transfected into C3H 10T1/2 cells and cells were collected after 2 d to examine: (A) EZH2 mRNA levels by RT-qPCR; (B) histone H3 K27 tri-methylation levels at the promoter region of Pref-1 gene by ChIP; (C) Pref-1 mRNA levels by RT-qPCR. These results are the averages of three independent experiments, and the error bars indicate standard deviations.
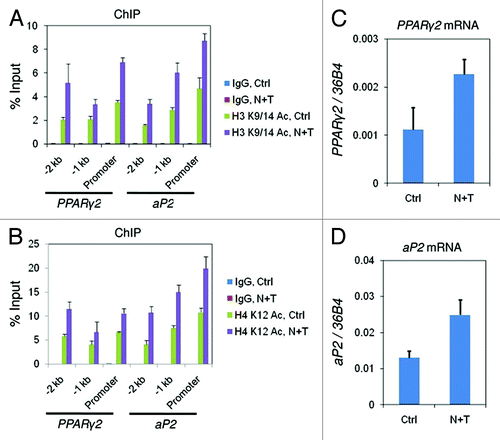
At the PPARγ2 and aP2 genes, we had found that both histone H3 and H4 acetylation positively correlate with gene expression during adipogenesis (; Figs. S1 and 7). To directly test whether the increase of histone acetylation affects the expression of these genes, we inhibited the HDACs by nicotinamide plus TSA treatment in 10T1/2 cells. The treatment lead to general increases (~2-fold) of both H3 K9/14 and H4 K12 acetylation at the promoter of PPARγ2 and aP2 genes (), as determined by ChIP assay. In parallel, we detected elevated expression of PPARγ2 and aP2 genes by RT-PCR ().
Figure 7. The increased levels of histone acetylation activate the transcription of PPARγ2 and aP2 genes. C3H 10T1/2 cells were treated with DMSO (Ctrl) or nicotinamide plus TSA (N + T) for 24 h before samples were collected for examination of: (A) histone H3 K9/14 acetylation and (B) histone H4 K12 acetylation levels at the promoter region of PPARγ2 and aP2 genes by ChIP; (C) PPARγ2 and (D) aP2 mRNA levels by RT-qPCR. These results are the averages of three independent experiments, and the error bars indicate standard deviations.
The active chromatin marker enriched 3′-UTR of C/EBPβ gene is a strong enhancer element
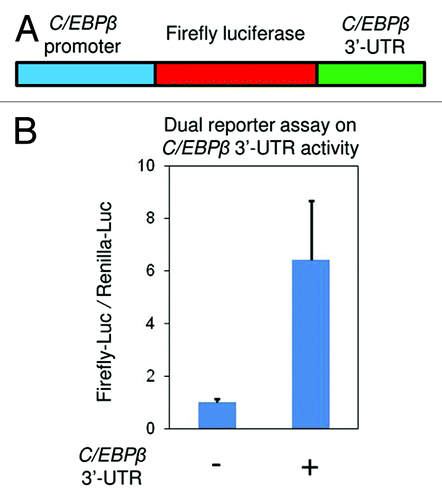
The intriguing co-occurrence of three active chromatin markers (, and ) at 3′-UTR of C/EBPβ gene promoted us to ask whether this DNA element plays a role in regulating C/EBPβ transcription. We used a dual luciferase reporter assay (Promega) to address this question (Experimental Procedures). We first inserted the C/EBPβ promoter upstream of firefly luciferase gene (), then examined the promoter activity in the presence or absence of the 3′-UTR sequence. As shown in , the conserved 3′-UTR significantly enhanced the C/EBPβ promoter activity by more than 6-folds, suggesting its essential role in the fast induction of C/EBPβ gene during the early stages of adipogenesis.
Figure 8. The active chromatin marker enriched 3′-UTR of C/EBPβ gene is a strong enhancer element. (A) The 3 kb C/EBPβ promoter was inserted upstream of firefly luciferase gene in pGL3 vector. The conserved 3′-UTR sequence was inserted downstream of the luciferase gene. (B) The firefly luciferase reporter vectors with or without C/EBPβ 3′-UTR were transfected into C3H 10T1/2 cells along with a renilla luciferase vector (pRLTK) as internal control. Activities of both firefly and renilla luciferases were measured 48 h after transfection. These results are the averages of three independent luciferase assays, and the error bars indicate standard deviations.
Discussion
Adipogenesis is regulated by a cascade of transcriptional events that require the concerted actions of various adipogenic factors. These factors include many histone-modifying enzymes,Citation18 suggesting that epigenetic mechanisms play essential roles in modulating adipogenesis. Through our systematic study of eight core histone modifications, we found that these modifications are stable at global levels but highly dynamic at the level of specific genes during adipogenesis, and their distribution patterns are distinct at different genes. Moreover, modulating these gene-specific histone modifications has a direct impact on the respective gene activity. These observations indicate that histone modifications have a well-defined “division of labor” in controlling the expression of various adipogenic regulatory genes.
Global levels of histone modifications
Genome-wide analyses of gene transcription during 3T3-L1 adipogenesis have shown that only a small fraction of the total genes are differentially expressed between preadipocytes and mature adipocytes.Citation40,Citation41 For example, Soukas et al. showed that among the 11,000 genes examined, only 1,259 genes changed their expression 3-fold or more during the course of differentiation. Half of these genes were upregulated, and the other half were downregulated.Citation40 In a later study, Burton et al. examined a total of 12,488 genes using expression microarrays and found that 1,686 transcripts showed a > 2-fold change during adipogenesis, with 859 transcripts increasing and 827 transcripts decreasing.Citation41 As histone modifications are tightly linked to gene transcription, we reasoned that the global levels of histone modifications are unlikely to show significant changes unless these modifications are predominantly enriched at groups of upregulated or downregulated genes, or there is an underlying cause unrelated to general gene expression change. Indeed, western blotting showed that the bulk levels of all eight examined histone modifications remain stable during the course of fat cell differentiation in both the 3T3-L1 and the 10T1/2 models (). Furthermore, these observations ruled out the possibility that the gene-specific changes in histone modification states are due to their fluctuation at the global level.
The Pref-1 gene
Pref-1 is highly expressed in 3T3-L1 preadipocytesCitation22 but was not detected in vivoCitation40 or in 10T1/2 MSCs before prolonged culture at confluence.Citation36 To investigate the epigenetic mechanism behind the differential expression of the Pref-1 gene in 3T3-L1 and 10T1/2 cells, we compared the distribution patterns of eight histone modifications between these two cell lines. Consistent with gene expression profiles (Fig. S1A), Pref-1 has lower levels of active chromatin markers, such as H3 K4 tri-methylation and H3 and H4 acetylation, in 10T1/2 cells than in 3T3-L1 cells (compare and ; Figs. S6C and 7). Significantly, we found a predominant enrichment of H3 K27 tri-methylation at the Pref-1 gene in 10T1/2 MSCs compared with the committed 3T3-L1 preadipocytes (compare ), suggesting that this repressive marker is involved in silencing the Pref-1 gene in 10T1/2 cells. Further functional study demonstrated that knocking down the K27 methylase Ezh2 leads to a decrease in K27 tri-methylation at the Pref-1 promoter and the release of Pref-1 repression in 10T1/2 cells (). This is consistent with a recent EZH2-knockout study.Citation31
The C/EBPβ gene
C/EBPβ is one of the factors induced during the early stages of adipogenesis, and it cooperates with C/EBPδ to activate downstream adipogenic regulators such as C/EBPα and PPARγ to fully establish the differentiation program.Citation42,Citation43 To investigate the epigenetic mechanism that controls the rapid activation of C/EBPβ upon induction (C/EBPβ usually reaches peak expression levels within a few hours), we examined the distribution of eight histone modifications at the C/EBPβ gene using ChIP. In 3T3-L1 cells, we found that the C/EBPβ gene is enriched for active chromatin markers such as H3 K4 tri-methylation, K9/K14 acetylation and H4 K20 mono-methylation, especially at its exon and conserved 3′-UTR (, and ). This conserved 3′-UTR was later shown as a strong enhancer element for C/EBPβ gene by luciferase assay (), suggesting that the histone modification patterns are useful resources for identification of important DNA elements for gene activity. In contrast to the enrichment of active chromatin markers, the entire C/EBPβ gene is devoid of repressive markers like H3 K9 and K27 tri-methylation (). Interestingly, the exon and 3′-UTR of the C/EBPβ gene only have baseline levels of K4 mono-methylation, allowing for hypermethylation of the same site by formation of tri-methyl groups. We also noticed that the acetylation patterns on histone H3 and H4 tails are completely different at the C/EBPβ gene, suggesting that these two histone tails play distinct roles in C/EBPβ regulation (). These observations were confirmed in 10T1/2 cells (Figs. S6–8). In human ES cells, key developmental genes are primed by the occupancy of both active and repressive histone markers,Citation13 but in preadipocytes and MSCs, we found that the C/EBPβ gene was marked only by active histone modifications. We interpret this observation to mean that the removal of repressive histone markers usually takes more than a few hours (as evidenced by the removal of H3 K27 tri-methylation from the C/EBPα locus, ), thus preventing the C/EBPβ gene from being rapidly induced and allowing the downstream adipogenic program to proceed in time. Therefore, the adipogenic precursor cells pre-mark this gene with only positive epigenetic markers and ready it for rapid induction. A recent reportCitation31 showed that abolishing H3 K27 tri-methylation by disrupting its KMT Ezh2 has no effect on C/EBPβ expression, and this observation further demonstrated that repressive histone markers are not involved in C/EBPβ regulation.
The C/EBPα gene
C/EBPα is one of the principle players in adipogenesis and is activated by C/EBPβ. Its essential roles in metabolism include maintaining the expression of PPARγ, controlling insulin activity and promoting white adipose tissue formation in vivo.Citation18 To gain insights into C/EBPα gene regulation, we profiled histone modification occupancies at this gene during the course of adipogenesis. We found that the C/EBPα gene is predominantly marked by H3 K27 tri-methylation in adipogenic precursor cells, and that this repressive marker decreases dramatically upon gene activation (). In addition, the C/EBPα gene is also enriched for H3 K4 tri-methylation (; Fig. S6C) during adipogenesis. These results indicate that the C/EBPα gene is under the control of bivalent histone markers in adipogenic precursor cells. Another repressive marker, H3 K9 tri-methylation, is enriched at the exon and conserved 3′-UTR of the C/EBPα gene but does not correlate with C/EBPα transcription (; Fig. S8A). The role of H3 K9 tri-methylation in C/EBPα gene regulation is currently unclear.
The PPARγ2 and aP2 genes
Numerous in vivo and in vitro data have established that PPARγ is the master regulator of adipogenesis.Citation19 PPARγ activates a whole range of protein factors to establish terminal adipogenesis. One of the targets of PPARγ is aP2, which is required for fatty acid binding and transportation in vivo. In this study, we observed that histone acetylation on both H3 and H4 tails positively correlates with transcription of the PPARγ2 and aP2 genes during adipogenesis (; Fig. S7). These results are consistent with a recent report showing that H3 K9 acetylation increases significantly at the PPARγ2 promoter during adipogenesis.Citation44 Our observation suggests that KATs play positive roles in regulating PPARγ targets, while HDACs are negative regulators of these genes. Indeed, previous reports showed that the KATs p300/CBPCitation34 and Tip60 (a MYST family member),Citation35 which catalyze histone H3 and H4 acetylation, respectively, bind to PPARγ and facilitate expression of its target genes. In contrast, SIRT1, a major HDAC in mammalian cells, binds to PPARγ and represses its targets.Citation33 Inhibiting HDACs by nicotinamide plus TSA treatment leads to increases of both H3 and H4 acetylation at the promoter of PPARγ2 and aP2 genes (), and this further leads to subsequent gene activation (). According to our ChIP assay, the PPARγ2 and aP2 genes are devoid of H3 K27 tri-methylation (), which coincides with the results of a recent genome-wide study.Citation45 These results indicate that this repressive histone marker and its corresponding modifying enzymes are not involved in regulating the expression of the PPARγ2 and aP2 genes.
In summary, our study has revealed distinct and highly dynamic patterns of histone modifications at different adipogenic regulatory genes. Modulation of these gene-specific histone modifications directly affects respective gene activity. Moreover, the histone modification patterns also serve as useful resources for predicting important DNA elements for gene regulation during adipogenesis. Future studies of chromatin modifications in other cell differentiation processes, such as myogenesis, should shed light on the epigenetic mechanisms controlling lineage commitment from progenitor cells under pathological conditions including aging.Citation46
Materials and Methods
Cell culture, differentiation and treatment
The mouse preadipocyte cell line 3T3-L1 and the mesenchymal stem cell line C3H 10T1/2 were purchased from the ATCC. The NIH 3T3 cell line was a kind gift from Dr. Xin-Yuan Fu (National University of Singapore). 3T3-L1 cells were maintained in DMEM containing 10% calf bovine serum (CBS) and differentiated in DMEM containing 10% fetal bovine serum (FBS), 1 µM DEX, 0.5 mM IBMX, and 10 µg/ml insulin for 2 d. Induction was continued for 4–6 d with DMEM containing 10% FBS and 10 µg/ml insulin. 10T1/2 cells were maintained in DMEM containing 10% FBS and differentiated by treatment with 1 µM DEX, 0.5 mM IBMX, 10 µg/ml insulin and 1 µM rosiglitazone (Rosi) for 2 d. This protocol was followed by treating the cells with 10 µg/ml insulin and 1 µM Rosi in DMEM containing 10% FBS for 4–6 d. NIH 3T3 cells were cultured in DMEM containing 10% CBS. EZH2 knockdown was performed in 10T1/2 cells using siGENOME SMARTpool siRNA targeting mouse EZH2 (14056) (Thermo Scientific). Cells were collected 48 h after siRNA transfection for further analyses. Nicotinamide plus Trichostatin A (TSA) treatment was performed in 10T1/2 cells at the final concentration of 10 mM nicotinamide and 100 nM TSA. Cells were treated for 24 h before they were collected for further analyses.
RNA extraction and RT-qPCR
Total RNA was extracted using TRIzol reagent (Invitrogen) at the indicated time points during the adipogenesis of 3T3-L1 and 10T1/2 cells. The RNA (1 µg) was treated with amplification grade DNase I (Invitrogen) and reverse transcribed with random 9-mer and M-MLV reverse transcriptase (Invitrogen). Expression of the key adipogenic regulatory genes Pref-1, C/EBPβ, C/EBPα, PPARγ2 and aP2 was determined by real-time quantitative PCR analysis using SYBR green and a 7900HT fast real-time PCR system (Applied Biosystems). Primer sequences for RT-qPCR are listed in Table S1. All gene expression data in this study were normalized to the riboprotein gene 36B4.Citation47
Histone purification and western blotting
Histone preparations from cells collected at different time points during adipogenesis were made as previously described.Citation48 Equal amounts of core histones were resolved by 15% SDS-PAGE, transferred onto PVDF membranes and probed with antibodies recognizing the following modifications: histone H3 K4 mono-methylation (Abcam, ab8895); H3 K4 di-methylation (Upstate, 07–030); H3 K4 tri-methylation (Upstate, 07–473); H3 K9/K14 acetylation (Upstate, 06–599); H4 K12 acetylation (Upstate, 07–595); H3 K9 tri-methylation (Abcam, ab8898); H3 K27 tri-methylation (Upstate, 07–449); H4 K20 mono-methylation (Abcam, ab9051) and total histone H3 (Abcam, ab1791).
Sequence conservation analysis
Sequence conservation analysis of the mouse adipogenic regulatory genes C/EBPα and C/EBPβ was conducted using the VISTA (visualization tool for alignment, www.genome.lbl.gov/vista/index.shtml) algorithm.Citation49 Mouse C/EBPα and C/EBPβ genomic sequences starting 2 kb upstream of the transcription start site (TSS) were compared with human and cow or human and rat sequences, respectively.
Chromatin immunoprecipitation
ChIP was performed essentially as described,Citation50 with the following modifications: cells were crosslinked with 1% formaldehyde for 10 min at 37°C, and then crude nuclei were purified using a previously described protocol.Citation48 Crude nuclei were sonicated using a Bioruptor UCD-300 (Diagenode) to obtain chromatin fragments approximately 500 bp in length. The antibodies used in the ChIP assay were the same as those described above for western blotting. Rabbit IgG (Sigma, I-5006) was included as a negative control. For each ChIP assay, 2–5 µg of antibodies was added and incubated overnight at 4°C. ChIP and input DNA were quantified by real-time quantitative PCR analysis using SYBR green and a 7900HT fast real-time PCR system (Applied Biosystems). ChIP-qPCR primers were designed to have melting temperatures (Tms) near 60°C. Primer Chr. Fifteen targets a silent region on mouse chromosome 15 containing no known genes within 500 kb.Citation35 Primer GAPDH targets the promoter region of the actively transcribed GAPDH gene. For all adipogenic regulator genes, -2 kb and -1 kb primers target the regions 2 kb and 1 kb upstream of the TSS, respectively. Promoter primers were designed to be within 500 bp upstream of the TSS for the gene in question. Large, conserved 3′-UTRs were found in both the C/EBPα and the C/EBPβ genes through sequence conservation analysis, and UTR primers were designed to target these regions. Primer sequences for ChIP-qPCR are listed in Table S2.
Plasmid construction and luciferase assay
A linker sequence, which contains six additional restriction sites: AatII, AgeI, ApaI, BssHII, NsiI and SpeI, was inserted into pGL3 basic vector (Promega) between the BamHI and SalI sites to yield plasmid pKR01. The 3 kb murine C/EBPβ promoterCitation51 was inserted upstream of firefly luciferase gene in pKR01 between the KpnI and XhoI sites to yield plasmid pKR02. A highly conserved 338-bp sequence from C/EBPβ 3′-UTR (+1167 to +1504 bp relative to TSS) was obtained through VISTA sequence conservation analysis and subsequently inserted downstream of the luciferase gene between the AatII and SalI sites in pKR02 to yield plasmid pKR03. Primer sequences for plasmid construction are listed in Table S3. Luciferase assay was performed by transfecting plasmid pKR02 or pKR03 into 10T1/2 cells along with a renilla luciferase vector (pRLTK) as internal control (Promega).
| Abbreviations: | ||
| MSC | = | mesenchymal stem cell |
| KAT | = | lysine acetyltransferase |
| HDAC | = | histone deacetylase |
| KMT | = | lysine methyltransferase |
| KDM | = | lysine demethylase |
| DEX | = | dexamethasone |
| IBMX | = | 3-isobutyl-1-methylxanthine |
| TSA | = | Trichostatin A |
| ChIP | = | chromatin immunoprecipitation |
| TSS | = | transcription start site |
Additional material
Download Zip (1.8 MB)Acknowledgments
The authors are grateful to the members of the Feng Xu laboratory for critical comments and discussion throughout this work. We are also grateful to Dr. Xin-Yuan Fu for the NIH 3T3 cell line. This work was supported by intramural funding from the Agency for Science, Technology and Research (A*STAR) to F.X.
References
- Kurdistani SK, Grunstein M. Histone acetylation and deacetylation in yeast. Nat Rev Mol Cell Biol 2003; 4:276 - 84; http://dx.doi.org/10.1038/nrm1075; PMID: 12671650
- Shogren-Knaak M, Ishii H, Sun JM, Pazin MJ, Davie JR, Peterson CL. Histone H4-K16 acetylation controls chromatin structure and protein interactions. Science 2006; 311:844 - 7; http://dx.doi.org/10.1126/science.1124000; PMID: 16469925
- Xu F, Zhang K, Grunstein M. Acetylation in histone H3 globular domain regulates gene expression in yeast. Cell 2005; 121:375 - 85; http://dx.doi.org/10.1016/j.cell.2005.03.011; PMID: 15882620
- Wysocka J, Swigut T, Milne TA, Dou Y, Zhang X, Burlingame AL, et al. WDR5 associates with histone H3 methylated at K4 and is essential for H3 K4 methylation and vertebrate development. Cell 2005; 121:859 - 72; http://dx.doi.org/10.1016/j.cell.2005.03.036; PMID: 15960974
- Jacobs SA, Taverna SD, Zhang Y, Briggs SD, Li J, Eissenberg JC, et al. Specificity of the HP1 chromo domain for the methylated N-terminus of histone H3. EMBO J 2001; 20:5232 - 41; http://dx.doi.org/10.1093/emboj/20.18.5232; PMID: 11566886
- Brownell JE, Zhou J, Ranalli T, Kobayashi R, Edmondson DG, Roth SY, et al. Tetrahymena histone acetyltransferase A: a homolog to yeast Gcn5p linking histone acetylation to gene activation. Cell 1996; 84:843 - 51; http://dx.doi.org/10.1016/S0092-8674(00)81063-6; PMID: 8601308
- Santos-Rosa H, Schneider R, Bannister AJ, Sherriff J, Bernstein BE, Emre NC, et al. Active genes are tri-methylated at K4 of histone H3. Nature 2002; 419:407 - 11; http://dx.doi.org/10.1038/nature01080; PMID: 12353038
- Rea S, Eisenhaber F, O’Carroll D, Strahl BD, Sun ZW, Schmid M, et al. Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature 2000; 406:593 - 9; http://dx.doi.org/10.1038/35020506; PMID: 10949293
- Ruthenburg AJ, Allis CD, Wysocka J. Methylation of lysine 4 on histone H3: intricacy of writing and reading a single epigenetic mark. Mol Cell 2007; 25:15 - 30; http://dx.doi.org/10.1016/j.molcel.2006.12.014; PMID: 17218268
- Wang Y, Jia S. Degrees make all the difference: the multifunctionality of histone H4 lysine 20 methylation. Epigenetics 2009; 4:273 - 6; http://dx.doi.org/10.4161/epi.4.5.9212; PMID: 19571682
- Heintzman ND, Stuart RK, Hon G, Fu Y, Ching CW, Hawkins RD, et al. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat Genet 2007; 39:311 - 8; http://dx.doi.org/10.1038/ng1966; PMID: 17277777
- Cao R, Wang L, Wang H, Xia L, Erdjument-Bromage H, Tempst P, et al. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science 2002; 298:1039 - 43; http://dx.doi.org/10.1126/science.1076997; PMID: 12351676
- Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell 2006; 125:315 - 26; http://dx.doi.org/10.1016/j.cell.2006.02.041; PMID: 16630819
- Allis CD, Berger SL, Cote J, Dent S, Jenuwien T, Kouzarides T, et al. New nomenclature for chromatin-modifying enzymes. Cell 2007; 131:633 - 6; http://dx.doi.org/10.1016/j.cell.2007.10.039; PMID: 18022353
- Kimura A, Matsubara K, Horikoshi M. A decade of histone acetylation: marking eukaryotic chromosomes with specific codes. J Biochem 2005; 138:647 - 62; http://dx.doi.org/10.1093/jb/mvi184; PMID: 16428293
- Shi Y, Lan F, Matson C, Mulligan P, Whetstine JR, Cole PA, et al. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell 2004; 119:941 - 53; http://dx.doi.org/10.1016/j.cell.2004.12.012; PMID: 15620353
- Klose RJ, Kallin EM, Zhang Y. JmjC-domain-containing proteins and histone demethylation. Nat Rev Genet 2006; 7:715 - 27; http://dx.doi.org/10.1038/nrg1945; PMID: 16983801
- Farmer SR. Transcriptional control of adipocyte formation. Cell Metab 2006; 4:263 - 73; http://dx.doi.org/10.1016/j.cmet.2006.07.001; PMID: 17011499
- Tontonoz P, Hu E, Spiegelman BM. Stimulation of adipogenesis in fibroblasts by PPAR gamma 2, a lipid-activated transcription factor. Cell 1994; 79:1147 - 56; http://dx.doi.org/10.1016/0092-8674(94)90006-X; PMID: 8001151
- Tontonoz P, Spiegelman BM. Fat and beyond: the diverse biology of PPARgamma. Annu Rev Biochem 2008; 77:289 - 312; http://dx.doi.org/10.1146/annurev.biochem.77.061307.091829; PMID: 18518822
- Ross SE, Hemati N, Longo KA, Bennett CN, Lucas PC, Erickson RL, et al. Inhibition of adipogenesis by Wnt signaling. Science 2000; 289:950 - 3; http://dx.doi.org/10.1126/science.289.5481.950; PMID: 10937998
- Smas CM, Sul HS. Pref-1, a protein containing EGF-like repeats, inhibits adipocyte differentiation. Cell 1993; 73:725 - 34; http://dx.doi.org/10.1016/0092-8674(93)90252-L; PMID: 8500166
- Wang Y, Sul HS. Pref-1 regulates mesenchymal cell commitment and differentiation through Sox9. Cell Metab 2009; 9:287 - 302; http://dx.doi.org/10.1016/j.cmet.2009.01.013; PMID: 19254573
- Cao Z, Umek RM, McKnight SL. Regulated expression of three C/EBP isoforms during adipose conversion of 3T3-L1 cells. Genes Dev 1991; 5:1538 - 52; http://dx.doi.org/10.1101/gad.5.9.1538; PMID: 1840554
- Yeh WC, Cao Z, Classon M, McKnight SL. Cascade regulation of terminal adipocyte differentiation by three members of the C/EBP family of leucine zipper proteins. Genes Dev 1995; 9:168 - 81; http://dx.doi.org/10.1101/gad.9.2.168; PMID: 7531665
- Tang QQ, Lane MD. Activation and centromeric localization of CCAAT/enhancer-binding proteins during the mitotic clonal expansion of adipocyte differentiation. Genes Dev 1999; 13:2231 - 41; http://dx.doi.org/10.1101/gad.13.17.2231; PMID: 10485846
- Musri MM, Corominola H, Casamitjana R, Gomis R, Párrizas M. Histone H3 lysine 4 dimethylation signals the transcriptional competence of the adiponectin promoter in preadipocytes. J Biol Chem 2006; 281:17180 - 8; http://dx.doi.org/10.1074/jbc.M601295200; PMID: 16613853
- Musri MM, Carmona MC, Hanzu FA, Kaliman P, Gomis R, Párrizas M. Histone demethylase LSD1 regulates adipogenesis. J Biol Chem 2010; 285:30034 - 41; http://dx.doi.org/10.1074/jbc.M110.151209; PMID: 20656681
- Lee J, Saha PK, Yang QH, Lee S, Park JY, Suh Y, et al. Targeted inactivation of MLL3 histone H3-Lys-4 methyltransferase activity in the mouse reveals vital roles for MLL3 in adipogenesis. Proc Natl Acad Sci USA 2008; 105:19229 - 34; http://dx.doi.org/10.1073/pnas.0810100105; PMID: 19047629
- Wakabayashi K, Okamura M, Tsutsumi S, Nishikawa NS, Tanaka T, Sakakibara I, et al. The peroxisome proliferator-activated receptor gamma/retinoid X receptor alpha heterodimer targets the histone modification enzyme PR-Set7/Setd8 gene and regulates adipogenesis through a positive feedback loop. Mol Cell Biol 2009; 29:3544 - 55; http://dx.doi.org/10.1128/MCB.01856-08; PMID: 19414603
- Wang L, Jin Q, Lee JE, Su IH, Ge K. Histone H3K27 methyltransferase Ezh2 represses Wnt genes to facilitate adipogenesis. Proc Natl Acad Sci USA 2010; 107:7317 - 22; http://dx.doi.org/10.1073/pnas.1000031107; PMID: 20368440
- Tateishi K, Okada Y, Kallin EM, Zhang Y. Role of Jhdm2a in regulating metabolic gene expression and obesity resistance. Nature 2009; 458:757 - 61; http://dx.doi.org/10.1038/nature07777; PMID: 19194461
- Picard F, Kurtev M, Chung N, Topark-Ngarm A, Senawong T, Machado De Oliveira R, et al. Sirt1 promotes fat mobilization in white adipocytes by repressing PPAR-gamma. Nature 2004; 429:771 - 6; http://dx.doi.org/10.1038/nature02583; PMID: 15175761
- Gelman L, Zhou G, Fajas L, Raspé E, Fruchart JC, Auwerx J. p300 interacts with the N- and C-terminal part of PPARgamma2 in a ligand-independent and -dependent manner, respectively. J Biol Chem 1999; 274:7681 - 8; http://dx.doi.org/10.1074/jbc.274.12.7681; PMID: 10075656
- van Beekum O, Brenkman AB, Grøntved L, Hamers N, van den Broek NJ, Berger R, et al. The adipogenic acetyltransferase Tip60 targets activation function 1 of peroxisome proliferator-activated receptor gamma. Endocrinology 2008; 149:1840 - 9; http://dx.doi.org/10.1210/en.2007-0977; PMID: 18096664
- Nueda ML, Baladrón V, García-Ramírez JJ, Sánchez-Solana B, Ruvira MD, Rivero S, et al. The novel gene EGFL9/Dlk2, highly homologous to Dlk1, functions as a modulator of adipogenesis. J Mol Biol 2007; 367:1270 - 80; http://dx.doi.org/10.1016/j.jmb.2006.10.020; PMID: 17320102
- Han M, Kim UJ, Kayne P, Grunstein M. Depletion of histone H4 and nucleosomes activates the PHO5 gene in Saccharomyces cerevisiae. EMBO J 1988; 7:2221 - 8; PMID: 3046934
- Reinke H, Hörz W. Histones are first hyperacetylated and then lose contact with the activated PHO5 promoter. Mol Cell 2003; 11:1599 - 607; http://dx.doi.org/10.1016/S1097-2765(03)00186-2; PMID: 12820972
- Boeger H, Griesenbeck J, Strattan JS, Kornberg RD. Nucleosomes unfold completely at a transcriptionally active promoter. Mol Cell 2003; 11:1587 - 98; http://dx.doi.org/10.1016/S1097-2765(03)00231-4; PMID: 12820971
- Soukas A, Socci ND, Saatkamp BD, Novelli S, Friedman JM. Distinct transcriptional profiles of adipogenesis in vivo and in vitro. J Biol Chem 2001; 276:34167 - 74; http://dx.doi.org/10.1074/jbc.M104421200; PMID: 11445576
- Burton GR, Nagarajan R, Peterson CA, McGehee RE Jr.. Microarray analysis of differentiation-specific gene expression during 3T3-L1 adipogenesis. Gene 2004; 329:167 - 85; http://dx.doi.org/10.1016/j.gene.2003.12.012; PMID: 15033539
- Lane MD, Tang QQ, Jiang MS. Role of the CCAAT enhancer binding proteins (C/EBPs) in adipocyte differentiation. Biochem Biophys Res Commun 1999; 266:677 - 83; http://dx.doi.org/10.1006/bbrc.1999.1885; PMID: 10603305
- Hamm JK, Park BH, Farmer SR. A role for C/EBPbeta in regulating peroxisome proliferator-activated receptor gamma activity during adipogenesis in 3T3-L1 preadipocytes. J Biol Chem 2001; 276:18464 - 71; http://dx.doi.org/10.1074/jbc.M100797200; PMID: 11279134
- Steger DJ, Grant GR, Schupp M, Tomaru T, Lefterova MI, Schug J, et al. Propagation of adipogenic signals through an epigenomic transition state. Genes Dev 2010; 24:1035 - 44; http://dx.doi.org/10.1101/gad.1907110; PMID: 20478996
- Mikkelsen TS, Xu Z, Zhang X, Wang L, Gimble JM, Lander ES, et al. Comparative epigenomic analysis of murine and human adipogenesis. Cell 2010; 143:156 - 69; http://dx.doi.org/10.1016/j.cell.2010.09.006; PMID: 20887899
- Nakamura K, Nakano S, Miyoshi T, Yamanouchi K, Matsuwaki T, Nishihara M. Age-related resistance of skeletal muscle-derived progenitor cells to SPARC may explain a shift from myogenesis to adipogenesis. Aging (Albany NY) 2012; 4:40 - 8; PMID: 22289652
- Laborda J. 36B4 cDNA used as an estradiol-independent mRNA control is the cDNA for human acidic ribosomal phosphoprotein PO. Nucleic Acids Res 1991; 19:3998; http://dx.doi.org/10.1093/nar/19.14.3998; PMID: 1861990
- Shechter D, Dormann HL, Allis CD, Hake SB. Extraction, purification and analysis of histones. Nat Protoc 2007; 2:1445 - 57; http://dx.doi.org/10.1038/nprot.2007.202; PMID: 17545981
- Dubchak I, Ryaboy DV. VISTA family of computational tools for comparative analysis of DNA sequences and whole genomes. Methods Mol Biol 2006; 338:69 - 89; PMID: 16888351
- Xie W, Song C, Young NL, Sperling AS, Xu F, Sridharan R, et al. Histone h3 lysine 56 acetylation is linked to the core transcriptional network in human embryonic stem cells. Mol Cell 2009; 33:417 - 27; http://dx.doi.org/10.1016/j.molcel.2009.02.004; PMID: 19250903
- Zhang K, Guo W, Yang Y, Wu J. JAK2/STAT3 pathway is involved in the early stage of adipogenesis through regulating C/EBPβ transcription. J Cell Biochem 2011; 112:488 - 97; http://dx.doi.org/10.1002/jcb.22936; PMID: 21268070