Regulated transcription requires transcription factors (TFs) and targeted response element sequences (REs). Variations in TF levels and sequence can “tune” a regulatory network. Although there is considerable information about REs for many TFs, relatively little is known about the influence of levels of TFs on transactivation. Given the importance of the human tumor suppressor p53 in many biological functions as well as in cancer, we wanted to understand transcriptional responses as a function of p53 level and target sequence, especially since the level of p53 in human cells can change many-fold in response to a variety of stress and cellular environments. The p53 levels could determine relative expression and, thus, biological consequences, such as cell cycle arrest and apoptosis.Citation1
We recently combined a yeast-based system that addresses p53 transactivation from many REs at various p53 levels,Citation2 with in vitro physical analyses of p53 interactions with the REs themselves to examine how the amount of p53 might influence transactivation.Citation3 Comparisons were made between in vivo transactivation and either the in vitro binding affinity (Kd) or the torsional flexibilities of the REs. At high levels of p53 in yeast, recapitulating p53 induction levels found after stress in human cells, there was a strong correlation between level of in vivo transactivation and in vitro binding affinity between 23 human-derived REs. This contrasts with transactivation at lower levels of p53, where we did not find a correlation between binding affinity and transactivation levels at the various REs, but instead found a strong correlation between DNA torsional flexibility and levels of transactivation of the 29 natural and synthetic target RE sequences analyzed.
We were guided into looking at DNA torsional flexibilities by our recent study,Citation4 where we discovered that changes in this parameter are the most significant difference between p53 REs. We were able to calculate it for any p53 RE by our observation that calculated DNA torsional flexibility, derived from crystal-structure analyses of protein-DNA complexes,Citation5 recapitulates the trend observed in the experimentally derived values.Citation4
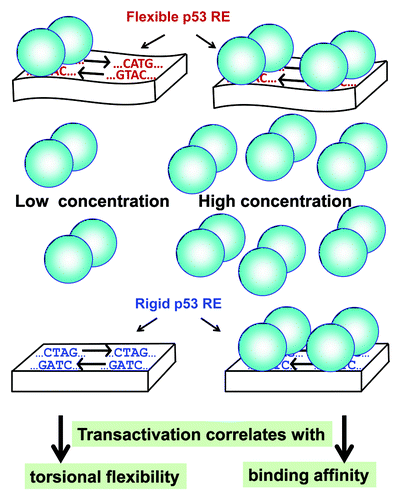
Since transactivation by p53 requires oligomerization of p53 to the functional tetramer we propose, as described in , that when the protein level is low, the chance that two dimeric p53 molecules (the prevalent species at low protein concentration)Citation6 arrive together at the RE to bind as a tetramer and provide transactivation is small. The RE must, therefore, provide kinetic stability to assure the stable binding of the first dimer until a second dimer comes along and interacts with the first one. Torsional flexibility of the RE can contribute to the kinetic stability of p53 dimers on the RE, because torsional flexibility of p53 REs can facilitate the reorientation of two p53 monomers within each dimer.Citation7 Thus, our findings establish that p53 transactivation is kinetically determined at low levels and thermodynamically driven at high levels.
Figure 1. Model of p53 bound to DNA at low (left) vs. high (right) p53 concentrations. p53 dimers are schematically represented by two connected balloons. The DNA RE is represented by a rectangular block when the DNA is rigid (containing the rigid CTAG motif) or by a wavy rectangular block when the DNA is flexible (containing the flexible CATG motif). The drawing is not to scale. The DNA double helix is narrower than the p53 spheres bound to it.
Among the many sequences examined, two artificial sequences (GGGCATGTCC x2, “Con-A” and GGGCATGCCC x2, “GGG”) had the unusual feature that they supported high levels of transactivation at very low (basal) levels of expression in both the yeast system (less than a few hundred molecules per cell) and when transfected into human cells. The transactivation response in yeast was at least 10-fold greater than from other highly responsive natural REs, such as the sequence associated with the p21/WAF1 gene, the classical p53 target generally considered to be among the most responsive. However, at high p53 levels, there was no difference in transactivation between the super-transactivating (STA) and the p21 target REs, suggesting a novel interaction with the STA target at low p53 concentrations. The STA phenomenon required that the sequences contain the flexible central motif CATG, have no spacer between adjacent half sites, and that p53 can form tetramers.
We suggest that the STA response is linked to the ability of p53 to form kinetically stable complexes with the STA sequences on torsional flexible targets. The off-rates for these REs was observed to be twice that for other flexible targets, such as the p21-5′ RE, which has a greater binding affinity to p53 than Con-A or GGG.Citation3
The STA sequences provide new opportunities to understand evolution of p53 REs, modification of p53 pathways, as well as analysis of p53 mutants. For example, there are no STAs in the human genome or any other genome examined, possibly suggesting potential problems of STAs at basal levels of p53, since there might be little control of transactivation. Given the high responsiveness of STAs, Con-A can be used to assess the potential of mutant p53s to function. About one-third of the tumor-associated p53 mutants retain transcription capability and can even result in a “change-of-spectrum” of the transactivated RE (IARC, version R15, November 2010, 18),Citation8 which can depend on p53 level. We found that several breast cancer-associated mutants could transactivate from Con-A, but not other well-established RE’s. The STA sequences might be useful for determining which p53 mutants could be remedied, suggesting general diagnostic and therapeutic utilities.
Overall, we establish that transactivation is determined by thermodynamics (i.e., binding affinity) at high p53 concentrations and by kinetic stability associated with RE flexibility at low levels. These findings have implications for other sequence-specific TFs that are subject to variable levels of control or variable levels of expression due to SNPs in REs, as well numerical changes in TFs, which might arise by chromosomal variation in copy number (i.e., CNVs).Citation9
References
- Resnick MA, et al. Proc Natl Acad Sci USA 2003; 100:9934 - 9; http://dx.doi.org/10.1073/pnas.1633803100; PMID: 12909720
- Jordan JJ, et al. PLoS Genet 2008; 4:e1000104; http://dx.doi.org/10.1371/journal.pgen.1000104; PMID: 18714371
- Jordan JJ, et al. Proc Natl Acad Sci USA 2012; 109:14387 - 92; http://dx.doi.org/10.1073/pnas.1205971109; PMID: 22908277
- Beno I, et al. Nucleic Acids Res 2011; 39:1919 - 32; http://dx.doi.org/10.1093/nar/gkq1044; PMID: 21071400
- Olson WK, et al. Proc Natl Acad Sci USA 1998; 95:11163 - 8; http://dx.doi.org/10.1073/pnas.95.19.11163; PMID: 9736707
- Rajagopalan S, et al. Nucleic Acids Res 2011; 39:2294 - 303; http://dx.doi.org/10.1093/nar/gkq800; PMID: 21097469
- Kitayner M, et al. Mol Cell 2006; 22:741 - 53; http://dx.doi.org/10.1016/j.molcel.2006.05.015; PMID: 16793544
- Petitjean A, et al. Hum Mutat 2007; 28:622 - 9; http://dx.doi.org/10.1002/humu.20495; PMID: 17311302
- Liu P, et al. Curr Opin Genet Dev 2012; 22:211 - 20; http://dx.doi.org/10.1016/j.gde.2012.02.012; PMID: 22440479