Abstract
Damage-specific DNA-binding protein 2 (DDB2) was first isolated as a subunit of the UV-DDB heterodimeric complex that is involved in DNA damage recognition in the nucleotide excision repair pathway (NER). DDB2 is required for efficient repair of CPDs in chromatin and is a component of the CRL4DDB2 E3 ligase that targets XPC, histones and DDB2 itself for ubiquitination. In this study, a yeast two-hybrid screening of a human cDNA library was performed to identify potential DDB2 cellular partners. We identified a deubiquitinating enzyme, USP24, as a likely DDB2-interacting partner. Interaction between DDB2 and USP24 was confirmed by co-precipitation. Importantly, knockdown of USP24 in two human cell lines decreased the steady-state levels of DDB2, indicating that USP24-mediated DDB2 deubiquitination prevents DDB2 degradation. In addition, we demonstrated that USP24 can cleave an ubiquitinated form of DDB2 in vitro. Taken together, our results suggest that the ubiquitin-specific protease USP24 is a novel regulator of DDB2 stability.
Introduction
Nucleotide excision repair (NER) is the principal pathway for repair of UV-induced cyclobutane pyrimidine dimers (CPDs) and (6-4) photoproducts in human cells. UV-DDB (UV-damaged DNA-binding protein) is a heterodimeric protein complex consisting of 127 kDa DDB1 and 48 kDa DDB2,Citation1-Citation4 which participates in damage recognition in global genome (GG)-NER. Mutations in DDB2 result in defective NER, as reflected in the heritable sun-sensitive skin condition, xeroderma pigmentosum group E (XP-E).Citation1,Citation4,Citation5 Cells derived from XP-E patients are deficient in CPD repair and show reduced 6–4PP repair.Citation2,Citation6-Citation9 Deletion of the DDB2 gene in mice significantly impairs the repair of UV lesions and leads to hypersensitivity to UV-induced skin cancers.Citation10
DDB2 is in complex with the CUL4-RING E3 ubiquitin ligase complex (CRL4DDB2), consisting of DDB1, CUL4 and RBX1.Citation11-Citation15 DDB2 is thought to be the substrate receptor targeting the E3 ligase complex to DNA damage sites to facilitate GG-NER. Of note, DDB1 and CUL4 have been shown to be in complex with other proteins, including CSA, a transcription-coupled, NER-specific protein.Citation11,Citation15 Consistent with its classification as an E3 ligase, XPC, histone H2A, H3, H4 and DDB2 itself have been identified as ubiquitination targets of the CRL4DDB2 E3 ligase complex.Citation12,Citation16-Citation22 The E3 ligase CRL4 DDB2 is found in complex with the COP9 signalosome (CSN).Citation15,Citation23-Citation25 In the absence of UV damage, CSN is associated with CRL4DDB2 and regulates its E3 ligase activity by deneddylation. After UV irradiation, CSN disassociates from CRL4DDB2, allowing DDB2 binding to the damage sites and subsequent DDB2 ubiquitination by CRL4DDB2.Citation24,Citation26
Several lines of evidence suggest that DDB2 plays a key role in the repair of UV damage only in the context of chromatin. Although UV-DDB binds strongly to UV damaged DNA,Citation27-Citation31 it stimulates NER of naked DNA only slightly in vitro.Citation32-Citation34 XP-E cell extracts display proficient NER of naked DNA in vitro, suggesting that UV-DDB has a role in the repair of DNA in chromatin.Citation35 DDB2 binds the lesion independent of XPC,Citation36 and XPC recruitment to UV damage is significantly decreased in the absence of functional DDB2.Citation10,Citation37,Citation38 DDB2 can co-localize with both CPDs and 6-4PPs in vivo, while XPC seems to bind 6-4PPs efficiently, but not CPDs. This suggests the necessity of DDB2 in GG-NER is specific for CPD repair.Citation38 Importantly, it has been suggested that the observed high affinity of DDB2 for 6-4PPs aids in the targeting of XPC to 6-4PPs when low levels of damage are present.Citation39 DDB2 autoubiquitination leads to the loss of DNA damage binding and rapid DDB2 degradation.Citation16,Citation19,Citation40-Citation42 XPC ubiquitination, in contrast, retains the complex at the site of UV damage without immediate proteasomal degradation. The differential response of XPC and DDB2 upon ubiquitination has been linked to an ubiquitin-dependent damage handover from DDB2 to XPC.Citation16,Citation43,Citation44 Recent findings show that UV-DDB associates preferentially with lesions in internucleosomal sites. In addition, UV-DDB and DDB2 ubiquitination are required to retain XPC at the linker regions. However, while UV-DDB facilitates XPC binding to nucleosomal DNA lesions, this does not appear to require DDB2 ubiquitination.Citation45 Luijsterburg et al., demonstrated that chromatin regions containing UV lesions undergo ATP-dependent chromatin decondensation that is strictly dependent on the presence of functional DDB2.Citation46 Incidentally, others and we have reported the association of UV-DDB with ATP-dependent chromatin remodeling factors.Citation47,Citation48 Furthermore, DDB2 has been shown to associate with the histone acetyltransferases CBP/p300.Citation41,Citation49 Clearly, understanding the role of DDB2 in NER will yield important insight into the mechanisms of NER operation in the context of chromatin.
Additionally, DDB2 has been implicated in an alternative process of the DNA damage response as well, via regulation of p21.Citation50-Citation52 DDB2 is involved in SOD2 transcription and stimulation of E2F1-dependent transcription targets.Citation53,Citation54 DDB2 has also been implicated in apoptosis due to a complex regulatory circuit between DDB2 and p53.Citation2,Citation55,Citation56 The ability of DDB2 to function in these processes potentially complicates the elucidation of mechanisms regulating its interaction with chromatin during NER. Therefore, identification of DDB2-interacting proteins will help elucidate the roles of DDB2 in not only DNA repair, but also other cellular processes. In this study, yeast two-hybrid screening of a human cDNA library allowed us to show that the ubiquitin-specific protease USP24 interacts with DDB2 and regulates DDB2 stability in human cells.
Results
Identify new DDB2-interacting proteins using yeast-two hybrid screening
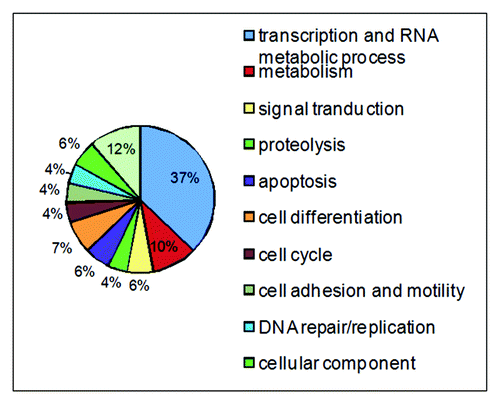
Besides its role in NER, DDB2 is involved in proteolysisCitation51 and transcriptional regulation.Citation57,Citation58 In order to gain a comprehensive picture of DDB2 functions, we took the yeast two-hybrid screen approach to establish a DDB2 protein interactome. We screened a normalized universal human cDNA library using the full-length DDB2 as the bait. In silico search of the NCBI database allowed the identification and classification of potential DDB2-interacting proteins into 11 diverse processes (). These processes include transcription and RNA metabolic process, protein degradation, metabolism, cell cycle, cell differentiation, apoptosis, DNA repair and signal transduction, cell adhesion and motility, cellular component. These data suggest that DDB2 may play previously unknown roles in many cellular processes and provide a framework for the understanding of DDB2 functions.
DDB2 interacts with USP24
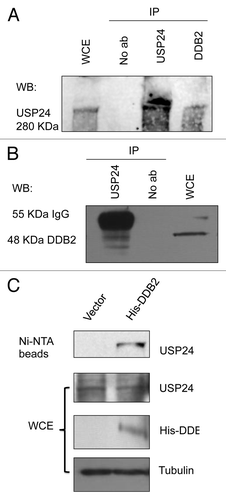
Among the potential DDB2-interacting proteins identified by the yeast-two hybrid screen, there are two ubiquitin-specific proteases (USPs), USP24 and USP53. USPs recognizes and removes the ubiquitin moiety from proteins.Citation59 The substrate of USP24 was previously unknown. To investigate whether DDB2 is a direct target of USP24, we set out to evaluate interactions in vivo by co-immunoprecipitation. By using the USP24-specific antibody, we examined the interaction between the endogenous DDB2 and USP24 proteins. Western blot analysis showed that USP24 was present in the anti-DDB2 immunoprecipitates from HeLa cell extracts, but not in the control immunoprecipitates (). Reciprocal co-immunoprecipitation experiment showed that DDB2 was detected in the anti-USP24 immunoprecipitates (). To avoid detecting the 55 KDa IgG heavy chains during western blotting, we expressed a His-tagged DDB2 in HeLa cells and precipitated His-DDB2 using Ni-NTA beads. Western blot showed again that USP24 was present in the His-DDB2 precipitates (). Importantly, USP24 was not present in the precipitates from extracts prepared from HeLa cells without His-DDB2 expression (). These observations suggest that USP24 interacts with DDB2 in vivo, as suggested by our yeast two-hybrid screen.
Figure 2. DDB2 interacts with USP24 in human cells. (A) Co-immunoprecipitation of USP24 and DDB2. IP was performed using an anti-DDB2 antibody, followed by western blot (WB) using an USP24 antibody. (B) Reciprocal co-immunoprecipitation of DDB2 and USP24. IP was performed using an anti-USP24 antibody, following by western blot (WB) using a DDB2 antibody. (C) His-tag pulldown of DDB2 precipitates USP24. His-tagged form of DDB2 was expressed in HeLa cells. Ni-NTA beads were used to pull down His-DDB2. HeLa cells without His-DDB2 expression were used as control to show specific DDB2 interaction with USP24.
USP24 regulates the stability of DDB2 in vivo
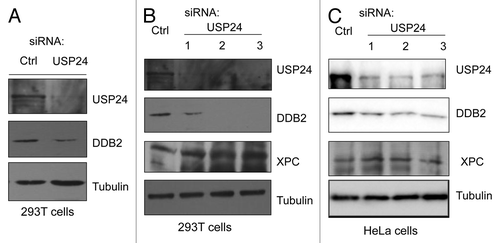
Ubiquitination is crucial in the physiological regulation of many cellular processes. DDB2 ubiquitination is mediated by the CRL4DDB2 E3 ligase, and its ubiquitination and subsequent degradation are essential for its functions in NER. USP24 belongs to the ubiquitin-specific processing protease family of deubiquitination enzymes (DUBs) and contains the characteristic Cys and His motifs at the core enzymatic domain.Citation59 It has the ability to remove ubiquitin from an ubiquitinated substrate, thereby regulating the stability and activity of this substrate. Since DDB2 is also ubiquitinated in human cells in the absence of exogenous DNA damage,Citation60 our identified interaction between DDB2 and USP24 immediately suggests a novel mechanism by which the ubiquitin moiety attached to DDB2 can be removed to prevent DDB2 degradation. To determine the functional relevance of DDB2-USP24 interactions, we tested whether USP24 affects the state-state levels of DDB2. As indicated in , knockdown of USP24 decreased the steady-state levels of DDB2 in both HeLa and 293T cells. Importantly, knockdown of USP24 using three different USP24-specific siRNAs all led to decreased levels of DDB2 in HeLa and 293T cells, when compared with the control siRNA-transfected cells (). In contrast, USP24 knockdown had no detectable effect on the levels of XPC in the two cell lines, another DNA lesion recognition protein which is also ubiquitinated by CL4DDB2, suggesting a specific role of USP24 in the control of DDB2 stability (). We note that, although UV irradiation induces the levels of ubiquitination of DDB2 and XPC,Citation16,Citation22 both proteins are ubiquitinated in human cells in the absence of exogenous DNA damage.Citation60
Figure 3. USP24 depletion in human cells destabilizes DDB2. (A) siRNA-mediated USP24 depletion in 293T cells decreases the steady-state levels of DDB2. (B) USP24 knockdown in 293T cells by three siRNAs destabilizes DDB2, but not XPC, in 293T cells. (C) Knockdown of USP24 in HeLa cells by three siRNAs and its effect on the steady-state levels of DDB2 and XPC.
USP53 interacts with DDB2, but knockdown of USP53 has no effect on DDB2 stability
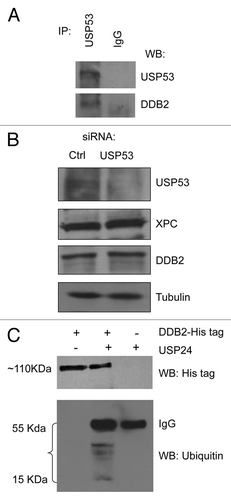
USP53 is another deubiquitinating enzyme identified in the yeast two-hybrid screening. Interestingly, we also confirmed that USP53 interacts with DDB2 by co-immunoprecipitation (). However, knockdown of USP53 expression in HeLa cells had no detectable effect on XPC and DDB2 expression (). Thus, the physiological relevance of USP53-DDB2 interactions is unclear.
Figure 4. (A) Co-immunoprecipitation of USP53 and DDB2. IP was performed using an anti-USP53 antibody, following by western blot (WB) using both USP53 and DDB2 antibodies. (B) Knockdown of USP53 in HeLa cells has no detectable effect on the steady-state levels of DDB2 and XPC. (C) Cleavage of ubiquitinated DDB2 in vitro by USP24. Ubiquitinated DDB2 was purified from UV-treated HeLa cells expressing Hig-tagged DDB2 in the presence of MG132. USP24 was partially purified from HeLa cells by immunoprecipitation using an USP24 antibody and protein G beads, followed by extensive washing to remove its binding partners. Cleavage reactions were performed at 37°C for 1 h.
USP24 cleaves an ubiquitinated form of DDB2 in vitro
To examine if USP24 can target ubiquitinated DDB2 in vitro, we treated HeLa cells expressing Hig-tagged DDB2 with MG132, a proteosome inhibitor,Citation61 for 1 h and exposed HeLa cells to UVC (10 J/m2) to stimulate DDB2 uniquitination for another hour, Ni-NTA beads were then used to pulldown modified His-DDB2. Using a His-tag-specific antibody, we detected a ~110 KDa band by western blot (Fig. 4C). Identical band could be picked up using an antibody specific for ubiquitin (data not shown), indicating that this is an ubiquitinated form of His-DDB2. Interestingly, addition of USP24 precipitated by an anti-USP24 antibody led to cleavage of this ubiquitinated form of His-DDB2, with the appearance of several bands corresponding to ubiquitin chains of different sizes (). Thus, USP24 can cleave an ubiquitinated form of DDB2 in vitro.
USP24, a nuclear protein, does not bind to UV damage sites
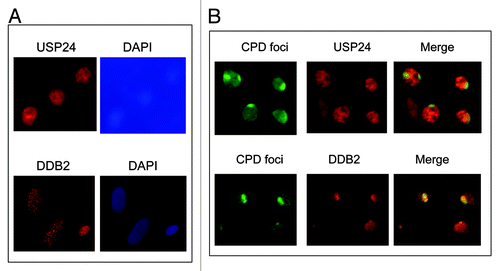
Since DDB2 is known to bind to UV damage sites in human cells and UV-stimulated DDB2 ubiquitination is believed to occur at sites of DNA damage, we next examined if USP24 also binds to UV damage sites. Immunofluorescence staining showed that both USP24 and DDB2 located primarily in the nucleus, suggesting that both are nuclear proteins (). However, unlike DDB2, which is recruited to CPD sites 15 min after UV irradiation (), USP24 did not accumulate at the CPD sites (). We also performed a time-course experiment but failed to detect USP24 accumulation at UV damage sites up to 3 h after UV irradiation (data not shown). These results suggest that USP24-mediated deubiquitination of modified DDB2 does not occur at the sites of DNA damage.
Figure 5. USP24, localized primarily in the nucleus, does not bind to UV damage sites. (A) Immunofluorescence staining of USP24 and DDB2. DAPI nucleus staining was shown. (B) Micropore UV irradiation followed by immunofluorescence staining using antibodies specific for USP24, UV lesion CPDs and DDB2.
Discussion
In this study, we identified a novel regulator of DDB2 stability using a yeast two-hybrid approach. We demonstrated biochemically that USP24 binds to DDB2 and regulates DDB2 stability in human cells. Our data suggest that USP24 stabilizes DDB2 by removing the ubiquitin moiety from modified DDB2, thereby preventing DDB2 degradation. As shown in , identified potential DDB2-interacting proteins largely fall into 11 classes, with some overlaps, on the basis of the cellular processes in which they are involved. These protein interaction data indicate that DDB2 may play other novel roles in human cells.
Key players in essential pathways are often subject to ubiquitin regulation mediated by one or several ubiquitin ligases or deubiquitinating enzymes (DUBs). Ubiquitin-specific proteases are a family of DUBs that participate in the removal of ubiquitin and ubiquitin-like modifications from an ubiquitinated substrate, thereby regulating the localization and activity of this substrate. Likewise, these proteases remove polyubiquitin from target proteins, rescuing them from degradation by the proteasome. The human genome encodes about 95 functional DUSs,Citation62 and only a few of those have been assigned functions or substrates. For example, USP7 has been shown to deubiquitinate p53, with overexpression of USP7 resulting in p53 stabilization.Citation63 In recent years, several USPs have been found to participate in the DNA damage response, including USP1,Citation64-Citation67 USP7,Citation68-Citation70 USP10,Citation71 USP13,Citation72 USP28Citation73 and USP29.Citation74 The function and substrate of USP24 were previously unknown. DDB2 represents the first example of a specific substrate that USP24 can directly deubiquitinate and stabilize in human cells.
Since DDB2 is a component of the CRL4DDB2 E3 ligase and one physiological substrate of this E3 ubiquitin ligase complex is DDB2 itself, it will be interesting to explore the regulation of DDB2 with or without DNA damage. Our results suggest that DDB2 modified by ubiquitination can be recycled by USP24 ( and ). We are investigating the involvement of USP24 in the control of DDB2 stability after UV irradiation. We speculate that there is a basal level of DDB2 ubiquitination in human cells, presumably mediated by the same CRL4DDB2 E3 ligase. Indeed, mass spectrometric analysis showed that DDB2 is also ubiquitinated in the cells in the absence of exogenous DNA damage.Citation60 Thus, USP24 plays an important role in the maintenance of steady-state levels of DDB2 by constantly removing the ubiquitin moiety from ubiquitinated DDB2, thereby preventing DDB2 degradation by the proteosomes ().
Figure 6. A model depicting the involvement of USP24 in DDB2 recycling. A low level of DDB2 is ubiquitinated continuously by the DDB1-CRL4DDB2 E3 ligase. We propose that USP24 deubiquitinates modified DDB2 and prevent its degradation by the proteasome. CSN-binding to DDB2 E3 ligase presumably inhibits DDB2 ubiquitination by silencing the stimulating effect of neddylation on the E3 ligase. Our yeast two-hybrid screen data suggest that interaction between the CRL4DDB2 E3 ligase and the CSN complex is bridged by the CSN2 subunit of CSN and DDB2.
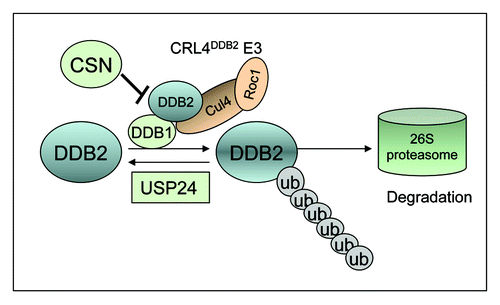
Interestingly, we identified COPS2/CSN2 from the yeast two-hybrid screen that may interact with DDB2 (data not shown). CSN2 is a subunit of CSN, an eight-subunit isopeptidase complex. CSN, via the proteolytic activity of its CSN5 subunit, removes the ubiquitin-like NEDD8 from cullins. Removal of NEDD8 consequently abrogates the stimulating effect of neddylation on cullin-based ubiquitin ligases.Citation75,Citation76 Recruitment of CRL4DDB2 to sites of DNA damage in chromatin appears to correlate with CSN release from the E3 ligase.Citation15,Citation24 Consistent with our data, CSN was co-immunoprecipitated with the DDB2 E3 complex from undamaged cells.Citation15 Our data further suggest that CSN2 may mediate the interaction between CSN complex and DDB2 E3 ligase to inhibit the basal level DDB2 ubiquitination (). Therefore, both CSN and USP24 are important for the control of the steady-state levels of DDB2: (1) CSN-binding to DDB2 E3 ligase presumably inhibits DDB2 ubiquitination by silencing the stimulating effect of neddylation on the E3 ligase; and (2) ubiquitinated DDB2 can be recycled by USP24 to prevent degradation ()
Materials and Methods
Yeast two-hybrid screen
Matchmaker Gold yeast two-hybrid screening was performed according to manufacturer’s instruction. Briefly, the full-length of the coding region of DDB2 was inserted in frame into the multiple cloning sites of the DNA-BD vector, pGBKT7 (Clontech), to generate the bait plasmid pGBKT7-DDB2, which was subsequently confirmed by sequencing. The pGBKT7-DDB2 was transformed into the bait strain Y2HGold. DDB2 bait strain Y2HGold was mated with the pre-transformed Y187/pACT2 normalized universal human Mate & Plate cDNA library according to the Clontech protocol. Diploid yeast cells were plated on a nutrient deficiency medium SD plate without Trp and Leu (DDO) and analyzed for their ability to grow in the presence of highly toxic drug Aureobasidin A (125 ng/ml, Clontech) and regulate α-galactosidase expression, which hydrolyzes 5-bromo-4-chloro-3-indolyl-a-d-galactopyranoside (X-α-gal, 40 µg/ml; Clontech) to produce a blue-end product. The selected colonies were restreaked on SD plate without Trp, Leu, His and Ade (QDO) containing Aureobasidin A and X-α-gal for further selection.
Sequencing and sequencing data analysis
Plasmid DNA from yeast was isolated and transformed into Escherichia coli DH5a for propagation. Plasmid DNA from E. coli was then sequenced (www.genewiz.com/). Nucleotide and deduced protein sequences were identified using BLAST (www.blast.ncbi.nlm.nih.gov/) and online services of EMBL-EBI (www.ebi.ac.uk/QuickGO/).
Cell culture
HeLa and 293T cells were obtained from ATCC. All cells were maintained in Dulbecco’s modified Eagle’s medium (Sigma) supplemented with 10% fetal bovine serum (Sigma) at 37°C and 5% CO2.
Co-immunoprecipitation
Cells from two 10-cm culture dishes were obtained and lysed with co-IP buffer (20 mM HEPES, 0.2 mM EDTA, 5% glycerol, 150 mM NaCl, 1% NP40, Roche complete EDTA-free protease inhibitor cocktail). Lysates were cleared by centrifugation (13,000 rpm, 10 min); the supernatant was incubated with USP24 antibody or DDB2 antibody overnight at 4°C and pulled down with protein G agarose beads (Millipore). Beads were washed three times with co-IP buffer and one time with TE buffer and eluted with one bead volume of 2X Laemmli SDS sample buffer at 95°C.
RNA interference
USP24 siRNA oligonucleotides and nonspecific siRNA were obtained from Sigma in a purified and annealed duplex form. The sequences, targeting three different regions of USP24 gene, were as follows: duplex-1, 5′-GAAACUCAGGGUUGAUACU-3′; duplex-2, 5′ CUUCUACUGUGGCUUGCUU-3′; duplex-3, 5′-GCACAAUACUGUGACCGUA-3′. siRNA transfections were performed with RNAiMAX (Invitrogen) according to the manufacturer’s instruction.
Antibodies, micropore UV irradiation and immunofluorescence staining
Protein concentrations of the cell lysates were quantified and separated by SDS-PAGE. Immunoblot analysis was performed using chemiluminescent detection. The antibodies were obtained from following sources: USP24 (1:1,000, Novus, Cat No: NB100–40830), DDB2 (1:500, Santa Cruz biotechnology, Cat No: sc-81246). Micropore UV irradiation and immunofluorescence staining were performed as described previously.Citation47
Acknowledgments
This work was supported by NIH R01 ES017784 and a summer undergraduate research supplement grant ES017784–3S1 from the National Institute of Environmental Health Sciences.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- Chu G, Chang E. Xeroderma pigmentosum group E cells lack a nuclear factor that binds to damaged DNA. Science 1988; 242:564 - 7; http://dx.doi.org/10.1126/science.3175673; PMID: 3175673
- Hwang BJ, Ford JM, Hanawalt PC, Chu G. Expression of the p48 xeroderma pigmentosum gene is p53-dependent and is involved in global genomic repair. Proc Natl Acad Sci USA 1999; 96:424 - 8; http://dx.doi.org/10.1073/pnas.96.2.424; PMID: 9892649
- Keeney S, Chang GJ, Linn S. Characterization of a human DNA damage binding protein implicated in xeroderma pigmentosum E. J Biol Chem 1993; 268:21293 - 300; PMID: 8407967
- Nichols AF, Ong P, Linn S. Mutations specific to the xeroderma pigmentosum group E Ddb- phenotype. J Biol Chem 1996; 271:24317 - 20; http://dx.doi.org/10.1074/jbc.271.40.24317; PMID: 8798680
- Tang J, Chu G. Xeroderma pigmentosum complementation group E and UV damaged DNA-binding protein. DNA Repair (Amst) 2002; 1:601 - 16; http://dx.doi.org/10.1016/S1568-7864(02)00052-6; PMID: 12509284
- Moser J, Volker M, Kool H, Alekseev S, Vrieling H, Yasui A, et al. The UV damaged DNA binding protein mediates efficient targeting of the nucleotide excision repair complex to UV-induced photo lesions. DNA Repair (Amst) 2005; 4:571 - 82; http://dx.doi.org/10.1016/j.dnarep.2005.01.001; PMID: 15811629
- Nichols AF, Itoh T, Graham JA, Liu W, Yamaizumi M, Linn S. Human damage-specific DNA-binding protein p48. Characterization of XPE mutations and regulation following UV irradiation. J Biol Chem 2000; 275:21422 - 8; http://dx.doi.org/10.1074/jbc.M000960200; PMID: 10777490
- Rapić-Otrin V, Navazza V, Nardo T, Botta E, McLenigan M, Bisi DC, et al. True XP group E patients have a defective UV damaged DNA binding protein complex and mutations in DDB2 which reveal the functional domains of its p48 product. Hum Mol Genet 2003; 12:1507 - 22; http://dx.doi.org/10.1093/hmg/ddg174; PMID: 12812979
- Tang JY, Hwang BJ, Ford JM, Hanawalt PC, Chu G. Xeroderma pigmentosum p48 gene enhances global genomic repair and suppresses UV-induced mutagenesis. Mol Cell 2000; 5:737 - 44; http://dx.doi.org/10.1016/S1097-2765(00)80252-X; PMID: 10882109
- Alekseev S, Kool H, Rebel H, Fousteri M, Moser J, Backendorf C, et al. Enhanced DDB2 expression protects mice from carcinogenic effects of chronic UV-B irradiation. Cancer Res 2005; 65:10298 - 306; http://dx.doi.org/10.1158/0008-5472.CAN-05-2295; PMID: 16288018
- Jackson S, Xiong Y. CRL4s: the CUL4-RING E3 ubiquitin ligases. Trends Biochem Sci 2009; 34:562 - 70; http://dx.doi.org/10.1016/j.tibs.2009.07.002; PMID: 19818632
- Kapetanaki MG, Guerrero-Santoro J, Bisi DC, Hsieh CL, Rapić-Otrin V, Levine AS. The DDB1-CUL4ADDB2 ubiquitin ligase is deficient in xeroderma pigmentosum group E and targets histone H2A at UV damaged DNA sites. Proc Natl Acad Sci USA 2006; 103:2588 - 93; http://dx.doi.org/10.1073/pnas.0511160103; PMID: 16473935
- Shiyanov P, Nag A, Raychaudhuri P. Cullin 4A associates with the UV damaged DNA-binding protein DDB. J Biol Chem 1999; 274:35309 - 12; http://dx.doi.org/10.1074/jbc.274.50.35309; PMID: 10585395
- He YJ, McCall CM, Hu J, Zeng Y, Xiong Y. DDB1 functions as a linker to recruit receptor WD40 proteins to CUL4-ROC1 ubiquitin ligases. Genes Dev 2006; 20:2949 - 54; http://dx.doi.org/10.1101/gad.1483206; PMID: 17079684
- Groisman R, Polanowska J, Kuraoka I, Sawada J, Saijo M, Drapkin R, et al. The ubiquitin ligase activity in the DDB2 and CSA complexes is differentially regulated by the COP9 signalosome in response to DNA damage. Cell 2003; 113:357 - 67; http://dx.doi.org/10.1016/S0092-8674(03)00316-7; PMID: 12732143
- Sugasawa K, Okuda Y, Saijo M, Nishi R, Matsuda N, Chu G, et al. UV-induced ubiquitylation of XPC protein mediated by UV-DDB-ubiquitin ligase complex. Cell 2005; 121:387 - 400; http://dx.doi.org/10.1016/j.cell.2005.02.035; PMID: 15882621
- Nag A, Bondar T, Shiv S, Raychaudhuri P. The xeroderma pigmentosum group E gene product DDB2 is a specific target of cullin 4A in mammalian cells. Mol Cell Biol 2001; 21:6738 - 47; http://dx.doi.org/10.1128/MCB.21.20.6738-6747.2001; PMID: 11564859
- Matsuda N, Azuma K, Saijo M, Iemura S, Hioki Y, Natsume T, et al. DDB2, the xeroderma pigmentosum group E gene product, is directly ubiquitylated by Cullin 4A-based ubiquitin ligase complex. DNA Repair (Amst) 2005; 4:537 - 45; http://dx.doi.org/10.1016/j.dnarep.2004.12.012; PMID: 15811626
- Chen X, Zhang Y, Douglas L, Zhou P. UV damaged DNA-binding proteins are targets of CUL-4A-mediated ubiquitination and degradation. J Biol Chem 2001; 276:48175 - 82; PMID: 11673459
- Wang H, Zhai L, Xu J, Joo HY, Jackson S, Erdjument-Bromage H, et al. Histone H3 and H4 ubiquitylation by the CUL4-DDB-ROC1 ubiquitin ligase facilitates cellular response to DNA damage. Mol Cell 2006; 22:383 - 94; http://dx.doi.org/10.1016/j.molcel.2006.03.035; PMID: 16678110
- Guerrero-Santoro J, Kapetanaki MG, Hsieh CL, Gorbachinsky I, Levine AS, Rapić-Otrin V. The cullin 4B-based UV damaged DNA-binding protein ligase binds to UV damaged chromatin and ubiquitinates histone H2A. Cancer Res 2008; 68:5014 - 22; http://dx.doi.org/10.1158/0008-5472.CAN-07-6162; PMID: 18593899
- Wang QE, Praetorius-Ibba M, Zhu Q, El-Mahdy MA, Wani G, Zhao Q, et al. Ubiquitylation-independent degradation of Xeroderma pigmentosum group C protein is required for efficient nucleotide excision repair. Nucleic Acids Res 2007; 35:5338 - 50; http://dx.doi.org/10.1093/nar/gkm550; PMID: 17693435
- Hannss R, Dubiel W. COP9 signalosome function in the DDR. FEBS Lett 2011; 585:2845 - 52; http://dx.doi.org/10.1016/j.febslet.2011.04.027; PMID: 21510940
- Takedachi A, Saijo M, Tanaka K. DDB2 complex-mediated ubiquitylation around DNA damage is oppositely regulated by XPC and Ku and contributes to the recruitment of XPA. Mol Cell Biol 2010; 30:2708 - 23; http://dx.doi.org/10.1128/MCB.01460-09; PMID: 20368362
- Olma MH, Roy M, Le Bihan T, Sumara I, Maerki S, Larsen B, et al. An interaction network of the mammalian COP9 signalosome identifies Dda1 as a core subunit of multiple Cul4-based E3 ligases. J Cell Sci 2009; 122:1035 - 44; http://dx.doi.org/10.1242/jcs.043539; PMID: 19295130
- Fischer ES, Scrima A, Böhm K, Matsumoto S, Lingaraju GM, Faty M, et al. The molecular basis of CRL4DDB2/CSA ubiquitin ligase architecture, targeting, and activation. Cell 2011; 147:1024 - 39; http://dx.doi.org/10.1016/j.cell.2011.10.035; PMID: 22118460
- Chu G, Yang W. Here comes the sun: recognition of UV damaged DNA. Cell 2008; 135:1172 - 4; http://dx.doi.org/10.1016/j.cell.2008.12.015; PMID: 19109889
- Scrima A, Konícková R, Czyzewski BK, Kawasaki Y, Jeffrey PD, Groisman R, et al. Structural basis of UV DNA damage recognition by the DDB1-DDB2 complex. Cell 2008; 135:1213 - 23; http://dx.doi.org/10.1016/j.cell.2008.10.045; PMID: 19109893
- Abramić M, Levine AS, Protić M. Purification of an ultraviolet-inducible, damage-specific DNA-binding protein from primate cells. J Biol Chem 1991; 266:22493 - 500; PMID: 1657999
- Fujiwara Y, Masutani C, Mizukoshi T, Kondo J, Hanaoka F, Iwai S. Characterization of DNA recognition by the human UV damaged DNA-binding protein. J Biol Chem 1999; 274:20027 - 33; http://dx.doi.org/10.1074/jbc.274.28.20027; PMID: 10391953
- Reardon JT, Nichols AF, Keeney S, Smith CA, Taylor JS, Linn S, et al. Comparative analysis of binding of human damaged DNA-binding protein (XPE) and Escherichia coli damage recognition protein (UvrA) to the major ultraviolet photoproducts: T[c,s]T, T[t,s]T, T[6-4]T, and T[Dewar]T. J Biol Chem 1993; 268:21301 - 8; PMID: 8407968
- Aboussekhra A, Biggerstaff M, Shivji MK, Vilpo JA, Moncollin V, Podust VN, et al. Mammalian DNA nucleotide excision repair reconstituted with purified protein components. Cell 1995; 80:859 - 68; http://dx.doi.org/10.1016/0092-8674(95)90289-9; PMID: 7697716
- Mu D, Park CH, Matsunaga T, Hsu DS, Reardon JT, Sancar A. Reconstitution of human DNA repair excision nuclease in a highly defined system. J Biol Chem 1995; 270:2415 - 8; http://dx.doi.org/10.1074/jbc.270.6.2415; PMID: 7852297
- Wakasugi M, Shimizu M, Morioka H, Linn S, Nikaido O, Matsunaga T. Damaged DNA-binding protein DDB stimulates the excision of cyclobutane pyrimidine dimers in vitro in concert with XPA and replication protein A. J Biol Chem 2001; 276:15434 - 40; http://dx.doi.org/10.1074/jbc.M011177200; PMID: 11278856
- Rapić Otrin V, Kuraoka I, Nardo T, McLenigan M, Eker AP, Stefanini M, et al. Relationship of the xeroderma pigmentosum group E DNA repair defect to the chromatin and DNA binding proteins UV-DDB and replication protein A. Mol Cell Biol 1998; 18:3182 - 90; PMID: 9584159
- Wakasugi M, Kawashima A, Morioka H, Linn S, Sancar A, Mori T, et al. DDB accumulates at DNA damage sites immediately after UV irradiation and directly stimulates nucleotide excision repair. J Biol Chem 2002; 277:1637 - 40; http://dx.doi.org/10.1074/jbc.C100610200; PMID: 11705987
- Wang QE, Zhu Q, Wani G, Chen J, Wani AA. UV radiation-induced XPC translocation within chromatin is mediated by damaged-DNA binding protein, DDB2. Carcinogenesis 2004; 25:1033 - 43; http://dx.doi.org/10.1093/carcin/bgh085; PMID: 14742321
- Fitch ME, Nakajima S, Yasui A, Ford JM. In vivo recruitment of XPC to UV-induced cyclobutane pyrimidine dimers by the DDB2 gene product. J Biol Chem 2003; 278:46906 - 10; http://dx.doi.org/10.1074/jbc.M307254200; PMID: 12944386
- Nishi R, Alekseev S, Dinant C, Hoogstraten D, Houtsmuller AB, Hoeijmakers JH, et al. UV-DDB-dependent regulation of nucleotide excision repair kinetics in living cells. DNA Repair (Amst) 2009; 8:767 - 76; http://dx.doi.org/10.1016/j.dnarep.2009.02.004; PMID: 19332393
- Fitch ME, Cross IV, Turner SJ, Adimoolam S, Lin CX, Williams KG, et al. The DDB2 nucleotide excision repair gene product p48 enhances global genomic repair in p53 deficient human fibroblasts. DNA Repair (Amst) 2003; 2:819 - 26; http://dx.doi.org/10.1016/S1568-7864(03)00066-1; PMID: 12826282
- Rapić-Otrin V, McLenigan MP, Bisi DC, Gonzalez M, Levine AS. Sequential binding of UV DNA damage binding factor and degradation of the p48 subunit as early events after UV irradiation. Nucleic Acids Res 2002; 30:2588 - 98; http://dx.doi.org/10.1093/nar/30.11.2588; PMID: 12034848
- Wittschieben BO, Iwai S, Wood RD. DDB1-DDB2 (xeroderma pigmentosum group E) protein complex recognizes a cyclobutane pyrimidine dimer, mismatches, apurinic/apyrimidinic sites, and compound lesions in DNA. J Biol Chem 2005; 280:39982 - 9; http://dx.doi.org/10.1074/jbc.M507854200; PMID: 16223728
- Sugasawa KUV-DDB. UV-DDB: a molecular machine linking DNA repair with ubiquitination. DNA Repair (Amst) 2009; 8:969 - 72; http://dx.doi.org/10.1016/j.dnarep.2009.05.001; PMID: 19493704
- El-Mahdy MA, Zhu Q, Wang QE, Wani G, Praetorius-Ibba M, Wani AA. Cullin 4A-mediated proteolysis of DDB2 protein at DNA damage sites regulates in vivo lesion recognition by XPC. J Biol Chem 2006; 281:13404 - 11; http://dx.doi.org/10.1074/jbc.M511834200; PMID: 16527807
- Fei J, Kaczmarek N, Luch A, Glas A, Carell T, Naegeli H. Regulation of nucleotide excision repair by UV-DDB: prioritization of damage recognition to internucleosomal DNA. PLoS Biol 2011; 9:e1001183; http://dx.doi.org/10.1371/journal.pbio.1001183; PMID: 22039351
- Luijsterburg MS, Lindh M, Acs K, Vrouwe MG, Pines A, van Attikum H, et al. DDB2 promotes chromatin decondensation at UV-induced DNA damage. J Cell Biol 2012; 197:267 - 81; http://dx.doi.org/10.1083/jcb.201106074; PMID: 22492724
- Zhang L, Zhang Q, Jones K, Patel M, Gong F. The chromatin remodeling factor BRG1 stimulates nucleotide excision repair by facilitating recruitment of XPC to sites of DNA damage. Cell Cycle 2009; 8:3953 - 9; http://dx.doi.org/10.4161/cc.8.23.10115; PMID: 19901545
- Jiang Y, Wang X, Bao S, Guo R, Johnson DG, Shen X, et al. INO80 chromatin remodeling complex promotes the removal of UV lesions by the nucleotide excision repair pathway. Proc Natl Acad Sci USA 2010; 107:17274 - 9; http://dx.doi.org/10.1073/pnas.1008388107; PMID: 20855601
- Datta A, Bagchi S, Nag A, Shiyanov P, Adami GR, Yoon T, et al. The p48 subunit of the damaged-DNA binding protein DDB associates with the CBP/p300 family of histone acetyltransferase. Mutat Res 2001; 486:89 - 97; http://dx.doi.org/10.1016/S0921-8777(01)00082-9; PMID: 11425514
- Stoyanova T, Yoon T, Kopanja D, Mokyr MB, Raychaudhuri P. The xeroderma pigmentosum group E gene product DDB2 activates nucleotide excision repair by regulating the level of p21Waf1/Cip1. Mol Cell Biol 2008; 28:177 - 87; http://dx.doi.org/10.1128/MCB.00880-07; PMID: 17967871
- Stoyanova T, Roy N, Kopanja D, Bagchi S, Raychaudhuri P. DDB2 decides cell fate following DNA damage. Proc Natl Acad Sci USA 2009; 106:10690 - 5; http://dx.doi.org/10.1073/pnas.0812254106; PMID: 19541625
- Bagchi S, Raychaudhuri P. Damaged-DNA Binding Protein-2 Drives Apoptosis Following DNA Damage. Cell Div 2010; 5:3; http://dx.doi.org/10.1186/1747-1028-5-3; PMID: 20205757
- Minig V, Kattan Z, van Beeumen J, Brunner E, Becuwe P. Identification of DDB2 protein as a transcriptional regulator of constitutive SOD2 gene expression in human breast cancer cells. J Biol Chem 2009; 284:14165 - 76; http://dx.doi.org/10.1074/jbc.M808208200; PMID: 19339246
- Hayes S, Shiyanov P, Chen X, Raychaudhuri P. DDB, a putative DNA repair protein, can function as a transcriptional partner of E2F1. Mol Cell Biol 1998; 18:240 - 9; PMID: 9418871
- Itoh T, Cado D, Kamide R, Linn S. DDB2 gene disruption leads to skin tumors and resistance to apoptosis after exposure to ultraviolet light but not a chemical carcinogen. Proc Natl Acad Sci USA 2004; 101:2052 - 7; http://dx.doi.org/10.1073/pnas.0306551101; PMID: 14769931
- Itoh T, O’Shea C, Linn S. Impaired regulation of tumor suppressor p53 caused by mutations in the xeroderma pigmentosum DDB2 gene: mutual regulatory interactions between p48(DDB2) and p53. Mol Cell Biol 2003; 23:7540 - 53; http://dx.doi.org/10.1128/MCB.23.21.7540-7553.2003; PMID: 14560002
- Ennen M, Minig V, Grandemange S, Touche N, Merlin JL, Besancenot V, et al. Regulation of the high basal expression of the manganese superoxide dismutase gene in aggressive breast cancer cells. Free Radic Biol Med 2011; 50:1771 - 9; http://dx.doi.org/10.1016/j.freeradbiomed.2011.03.013; PMID: 21419216
- Minig V, Kattan Z, van Beeumen J, Brunner E, Becuwe P. Identification of DDB2 protein as a transcriptional regulator of constitutive SOD2 gene expression in human breast cancer cells. J Biol Chem 2009; 284:14165 - 76; http://dx.doi.org/10.1074/jbc.M808208200; PMID: 19339246
- Quesada V, Díaz-Perales A, Gutiérrez-Fernández A, Garabaya C, Cal S, López-Otín C. Cloning and enzymatic analysis of 22 novel human ubiquitin-specific proteases. Biochem Biophys Res Commun 2004; 314:54 - 62; http://dx.doi.org/10.1016/j.bbrc.2003.12.050; PMID: 14715245
- Wagner SA, Beli P, Weinert BT, Nielsen ML, Cox J, Mann M, et al. A proteome-wide, quantitative survey of in vivo ubiquitylation sites reveals widespread regulatory roles. Mol Cell Proteomics 2011; 10:M111 - , 013284; http://dx.doi.org/10.1074/mcp.M111.013284; PMID: 21890473
- Zhang L, Hu JJ, Gong F. MG132 inhibition of proteasome blocks apoptosis induced by severe DNA damage. Cell Cycle 2011; 10:3515 - 8; http://dx.doi.org/10.4161/cc.10.20.17789; PMID: 22031102
- Sowa ME, Bennett EJ, Gygi SP, Harper JW. Defining the human deubiquitinating enzyme interaction landscape. Cell 2009; 138:389 - 403; http://dx.doi.org/10.1016/j.cell.2009.04.042; PMID: 19615732
- Li M, Chen D, Shiloh A, Luo J, Nikolaev AY, Qin J, et al. Deubiquitination of p53 by HAUSP is an important pathway for p53 stabilization. Nature 2002; 416:648 - 53; http://dx.doi.org/10.1038/nature737; PMID: 11923872
- Williams SA, Maecker HL, French DM, Liu J, Gregg A, Silverstein LB, et al. USP1 deubiquitinates ID proteins to preserve a mesenchymal stem cell program in osteosarcoma. Cell 2011; 146:918 - 30; http://dx.doi.org/10.1016/j.cell.2011.07.040; PMID: 21925315
- Jones MJ, Colnaghi L, Huang TT. Dysregulation of DNA polymerase κ recruitment to replication forks results in genomic instability. EMBO J 2012; 31:908 - 18; http://dx.doi.org/10.1038/emboj.2011.457; PMID: 22157819
- Cotto-Rios XM, Jones MJ, Busino L, Pagano M, Huang TT. APC/CCdh1-dependent proteolysis of USP1 regulates the response to UV-mediated DNA damage. J Cell Biol 2011; 194:177 - 86; http://dx.doi.org/10.1083/jcb.201101062; PMID: 21768287
- Oestergaard VH, Langevin F, Kuiken HJ, Pace P, Niedzwiedz W, Simpson LJ, et al. Deubiquitination of FANCD2 is required for DNA crosslink repair. Mol Cell 2007; 28:798 - 809; http://dx.doi.org/10.1016/j.molcel.2007.09.020; PMID: 18082605
- Zhang X, Horibata K, Saijo M, Ishigami C, Ukai A, Kanno SI, et al. Mutations in UVSSA cause UV-sensitive syndrome and destabilize ERCC6 in transcription-coupled DNA repair. Nat Genet 2012; 44:593 - 7; http://dx.doi.org/10.1038/ng.2228; PMID: 22466612
- Khoronenkova SV, Dianova II, Ternette N, Kessler BM, Parsons JL, Dianov GL. ATM-dependent downregulation of USP7/HAUSP by PPM1G activates p53 response to DNA damage. Mol Cell 2012; 45:801 - 13; http://dx.doi.org/10.1016/j.molcel.2012.01.021; PMID: 22361354
- Schwertman P, Lagarou A, Dekkers DH, Raams A, van der Hoek AC, Laffeber C, et al. UV-sensitive syndrome protein UVSSA recruits USP7 to regulate transcription-coupled repair. Nat Genet 2012; 44:598 - 602; http://dx.doi.org/10.1038/ng.2230; PMID: 22466611
- Yuan J, Luo K, Zhang L, Cheville JC, Lou Z. USP10 regulates p53 localization and stability by deubiquitinating p53. Cell 2010; 140:384 - 96; http://dx.doi.org/10.1016/j.cell.2009.12.032; PMID: 20096447
- Zhao X, Fiske B, Kawakami A, Li J, Fisher DE. Regulation of MITF stability by the USP13 deubiquitinase. Nat Commun 2011; 2:414; http://dx.doi.org/10.1038/ncomms1421; PMID: 21811243
- Zhang D, Zaugg K, Mak TW, Elledge SJ. A role for the deubiquitinating enzyme USP28 in control of the DNA damage response. Cell 2006; 126:529 - 42; http://dx.doi.org/10.1016/j.cell.2006.06.039; PMID: 16901786
- Liu J, Chung HJ, Vogt M, Jin Y, Malide D, He L, et al. JTV1 co-activates FBP to induce USP29 transcription and stabilize p53 in response to oxidative stress. EMBO J 2011; 30:846 - 58; http://dx.doi.org/10.1038/emboj.2011.11; PMID: 21285945
- Cope GA, Suh GS, Aravind L, Schwarz SE, Zipursky SL, Koonin EV, et al. Role of predicted metalloprotease motif of Jab1/Csn5 in cleavage of Nedd8 from Cul1. Science 2002; 298:608 - 11; http://dx.doi.org/10.1126/science.1075901; PMID: 12183637
- Lyapina S, Cope G, Shevchenko A, Serino G, Tsuge T, Zhou C, et al. Promotion of NEDD-CUL1 conjugate cleavage by COP9 signalosome. Science 2001; 292:1382 - 5; http://dx.doi.org/10.1126/science.1059780; PMID: 11337588