Abstract
Cyclins control cell cycle progression by regulating the activity of cyclin-dependent kinases (Cdks). Cyclin I is a member of the cyclin family because of the presence of a cyclin box motif. It has been suggested that Cyclin I is involved in various biological processes, such as cell survival, angiogenesis, and cell differentiation. However, whether or not Cyclin I has a role in regulating the cell cycle similarly to other cyclins has yet to be clarified. Therefore, we investigated the role for Cyclin I in cell cycle progression. We showed that the protein level of Cyclin I oscillated during the cell cycle, and that Cyclin I was subjected to ubiquitination and degradation in cells. The interaction between Cyclin I and Cdk5 was detected in cells overexpressed with both proteins. Furthermore, depletion of Cyclin I by siRNAs prevented cell proliferation, suggesting the positive role of Cyclin I for the cell cycle progression. In addition, flow cytometric analysis revealed that cells depleted of Cyclin I were accumulated at G2/M phases. By using HeLa.S-Fucci (fluorescent ubiquitination-based cell cycle indicator) cells, we further confirmed that knockdown of Cyclin I induced cell cycle arrest at S/G2/M phases. These results strongly suggest that Cyclin I has the role in the regulation of cell cycle progression.
Introduction
Cyclins are the regulatory subunits of cyclin-dependent kinases (Cdks), and the complexes of cyclin and Cdk play key roles in the control of cell cycle progression.Citation1-Citation3 Cyclins contain a well-conserved amino acid sequence known as the cyclin box, which is required for the binding to and activation of specific target Cdks in each cell cycle phase. Although levels of Cdks remain constant throughout the cell cycle, the activitiy of Cdks oscillate due to cell cycle-dependent phosphorylation and changes in the amounts of cyclins. In addition, the ubiquitin-mediated degradation of cyclins is critical for proper cell cycle progression.
Cyclin I was originally cloned from the human forebrain cortex.Citation4 It contains a typical cyclin box near the N terminus and a PEST-rich region near the C terminus and shows the highest sequence similarity to Cyclins G1 and G2. In contrast to other cyclins, the Cyclin I mRNA level does not fluctuate during cell cycle progression.Citation4-Citation6 Although the partner Cdk had not been identified for a long time, it has recently been reported that Cyclin I binds to and activates Cdk5, thereby preventing apoptosis in podocytes.Citation6 While most Cdks are activated in proliferating cells to promote cell cycle progression, the Cdk5 activity is predominantly detected in post-mitotic neurons,Citation7 and Cdk5 plays a role in a variety of neuronal functions, such as neuronal development, migration, and synaptic signaling.Citation8
Recent studies have suggested that Cyclin I expression is correlated with the proliferative activity and angiogenesis in human cancers,Citation9,Citation10 and that increased levels of Cyclin I are associated with terminal growth arrest in cardiomyocytes.Citation11 However, it is unclear whether Cyclin I has a direct role in regulating cell proliferation, similar to other cyclins. Therefore, we investigated the function of Cyclin I in the regulation of cell cycle progression. We report here that the protein level of Cyclin I increased during S phase, and that Cyclin I was ubiquitinated and degraded by the proteasome in cells. Furthermore, knockdown of Cyclin I prevented cell proliferation through the cell cycle arrest at S and G2/M phases. These results suggest that Cyclin I is involved in cell cycle progression.
Results
The protein level of Cyclin I oscillates during the cell cycle
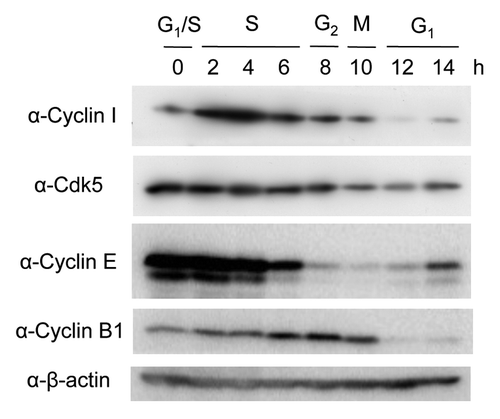
It has been suggested that the levels of many proteins involved in the cell cycle regulation are controlled by the ubiquitin-proteasome system.Citation1-Citation3 Although it was reported that the Cyclin I mRNA level did not change during cell cycle progression,Citation4-Citation6 the protein level of Cyclin I during the cell cycle remains unclear. Therefore, we examined whether Cyclin I was regulated during the cell cycle at the protein level. After synchronization of HeLa cells at late G1 phase with a double-thymidine block, followed by the release into the cell cycle, cells were collected at various time points and analyzed by immunoblotting (). The cells harvested at the indicated times were classified into respective cell cycle phase according to the protein levels of Cyclin E as a G1/S-phase marker and Cyclin B1 as a G2/M-phase marker. Although the level of Cyclin I was low at the border of G1 and S phases, the protein was upregulated during S phase and gradually decreased during G2, M, and G1 phases. As it has been reported that Cdk5 is a binding partner of Cyclin I and promotes cell survival,Citation12 we analyzed the protein level of Cdk5 during cell cycle progression. The Cdk5 protein was constantly detected during G1, S, and G2 phases, while the level of Cdk5 was slightly decreased during M and early G1 phases, indicating that Cdk5 largely unchanged with cell cycle. These results suggested that the protein level of Cyclin I oscillates during cell cycle progression, partially overlapping with Cdk5, in particular at S phase.
Figure 1. The protein level of Cyclin I oscillates during the cell cycle. HeLa cells were arrested at late G1 phase with the double-thymidine block, followed by the release to enter S phase. Lysates from cells at each time point were subjected to SDS-PAGE, followed by immunoblotting with antibodies as indicated. Cyclin E and Cyclin B1 were used for the cell cycle markers of G1/S and G2/M phases, respectively.
Cyclin I is degraded via the ubiquitin-proteasome pathway
Cyclin I contains the C-terminal PEST motif (amino acids 280–364)Citation4 known as a destabilization domain,Citation13 and the protein level of Cyclin I decreased at M and early G1 phases (). Most proteins with a very short half-life, including cyclins, are degraded by the ubiquitin-proteasome system.Citation14,Citation15 Therefore, we tested if Cyclin I is degraded via the ubiquitin-proteasome pathway. HEK293T cells were transiently transfected with pDsRed-Cyclin I and treated with or without proteasome inhibitor, lactacystin, for 24 h (). DsRed-Cyclin I-positive cells were apparently increased by the treatment with lactacystin, while the ratio of DsRed-positive cells in pDsRed transfection were not affected despite the presence of lactacystin. Furthermore, immunoblot analysis confirmed that the DsRed-Cyclin I protein level increased by the inhibition of proteasome (), suggesting that Cyclin I is degraded via the proteasome pathway.
Figure 2. Cyclin I is degraded via the ubiquitin-proteasome pathway. (A and B) Increasing level of Cyclin I by proteasome inhibition. HEK293T cells were transfected with pDsRed-Cyclin I. After incubation for 16 h, cells were treated with or without 10 μg/ml lactacystin for 24 h. Cells were observed with fluorescence microscope (A) or the lysates were fractionated by SDS-PAGE and immunoblotted with antibodies as indicated (B). Bars, 50 μm. (C) Ubiquitination of Cyclin I. HEK293T cells were transfected with pTB701-Flag-Cyclin I-Wt, pTB701-Flag-Cyclin I-ΔPEST, and pCGN-HA-ubiquitin as indicated. After incubation for 24 h, cells were treated with 10 μg/ml lactacystin for 6 h, and the lysates were immunoprecipitated with anti-Flag M2 agarose beads. The input lysates and the immunoprecipitates were fractionated by SDS-PAGE and immunoblotted with antibodies as indicated.
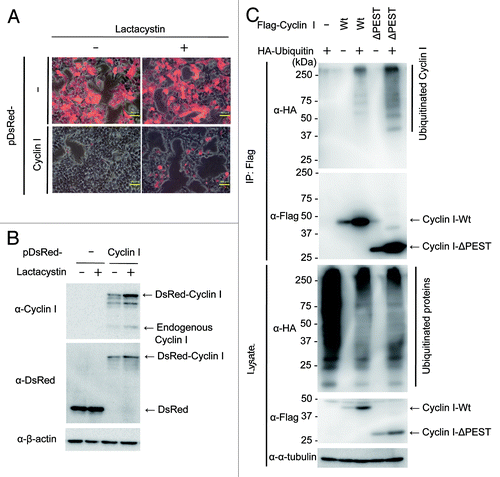
To determine whether Cyclin I is ubiquitinated, Flag-tagged Cyclin I was transiently overexpressed in HEK293T cells together with hemagglutinin (HA)-tagged ubiquitin (). After immunoprecipitation of Flag-Cyclin I, ubiquitinated Cyclin I was detected with anti-HA antibodies at high molecular weight regions when coexpressed with HA-ubiquitin, suggesting that Cyclin I is ubiquitinated in cells. To further investigate the involvement of the PEST-rich region on Cyclin I ubiquitination, we generated a PEST motif deletion mutant (Cyclin I-ΔPEST; amino acids 1–279). When immunoprecipitated, ubiquitinated Cyclin I-ΔPEST was also detected in the presence of HA-ubiquitin (). Although the level of ubiquitinated Cyclin I-ΔPEST appeared to be higher than that of ubiquitinated Cyclin I-Wt, given that Cyclin I-ΔPEST was more efficiently immunoprecipitated than Cyclin I-Wt, the ratio of the ubiquitinated form to the unmodified form was unchanged between Cyclin I-Wt and Cyclin I-ΔPEST. These results suggested that PEST-deleted Cyclin I is ubiquitinated similarly to full-length Cyclin I, and that the PEST-rich region is not involved in the degradation of Cyclin I.
Cyclin I associates with Cdk5 in vivo
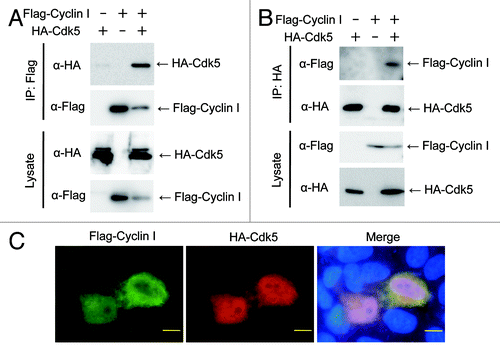
Next we tested whether an in vivo association between Cyclin I and Cdk5 could be detected by coimmunoprecipitation (). Flag-Cyclin I was transiently overexpressed in HEK293T cells together with HA-tagged Cdk5. After immunoprecipitation of Flag-Cyclin I, coprecipitated HA-Cdk5 was detected with anti-HA antibody (). Additionally, Flag-Cyclin I was also coprecipitated with HA-Cdk5 (). These results suggested that Cyclin I interacts with Cdk5 in vivo. To confirm whether Cyclin I is colocalized with Cdk5 in cells, HeLa cells transiently overexpressed with Flag-Cyclin I and HA-Cdk5 were stained with anti-Cyclin I and anti-HA antibodies (). Both of Cyclin I and Cdk5 were distributed throughout the cell, with a weak tendency for the enhanced signal of Cyclin I to be observed in the cytoplasm. Although both of Cyclin I and Cdk5 were detected throughout the cell, the signals of Cyclin I and Cdk5 were not completely overlapping even though both proteins expressed in the same cell. These data suggested that Cyclin I can function in part mediated through complex formation with Cdk5.
Figure 3. Cyclin I associates with Cdk5 in vivo. (A and B) Coprecipitation of Cyclin I with Cdk5 in transiently overexpressed HEK293T cells. Lysates from HEK293T cells transfected with pTB701-Flag-Cyclin I and pcDNA3-HA-Cdk5 were immunoprecipitated with anti-Flag agarose beads (A) or anti-HA antibody (B). The input lysates and the immunoprecipitates were fractionated by SDS-PAGE and immunoblotted with antibodies as indicated. (C) Colocalization of Cyclin I and Cdk5 in transiently overexpressed HeLa cells. HeLa cells were transfected with pTB701-Flag-Cyclin I and pcDNA3-HA-Cdk5. After fixation and permeabilization, cells were incubated with anti-Cyclin I antibody and anti-HA antibody, followed by staining nuclei with Hoechst 33258. Bars, 10 μm.
Knockdown of Cyclin I inhibits cell cycle progression
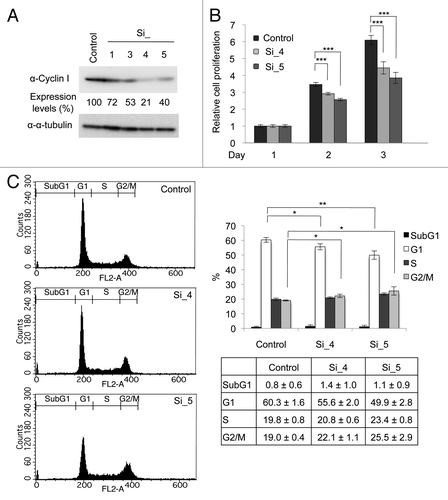
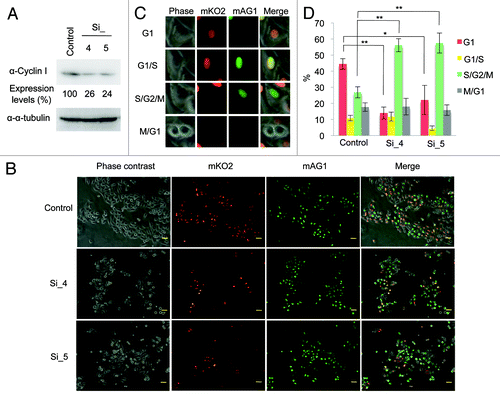
To investigate the role of Cyclin I in cell cycle progression, we used siRNAs to deplete Cyclin I (). HeLa cells were transfected with siRNAs for Cyclin I (Si_1, Si_3, Si_4, and Si_5) twice. Two days after the second transfection of siRNAs, the level of Cyclin I was decreased, whereas the level of α-tubulin was unaffected (). Because Cyclin I decreased particularly in cells transfected with Si_4 or Si_5, these siRNAs were used in subsequent experiments. Proliferation of cells transfected with siRNAs was monitored by WST-1 assay (). Knockdown of Cyclin I effectively prevented cell proliferation, suggesting that Cyclin I contributes to cell proliferation. To further confirm the effects of Cyclin I knockdown in cell cycle progression, cell cycle profiles were determined by flow cytometry (). When Cyclin I was depleted, cells at G2/M phases slightly increased, accompanying a decrease of cells at G1 phase, suggesting that Cyclin I plays the role in cell cycle progression around G2/M phases.
Figure 4. Knockdown of Cyclin I inhibits cell cycle progression in HeLa cells. (A) Knockdown of Cyclin I in HeLa cells. HeLa cells were transfected with siRNAs for negative control (control) or Cyclin I (Si_1, Si_3, Si_4, and Si_5) at day 0 and day 1, and then the expression level of Cyclin I was determined by immunoblot analysis at day 3. The protein level relative to the α-tubulin level was quantified using NIH ImageJ software. (B) The proliferation rates of Cyclin I-depleted HeLa cells. HeLa cells were cultured as described in (A), and cell proliferation was assessed by WST-1 assay at days 1, 2, and 3. (C) Cell cycle profiles of Cyclin I-depleted HeLa cells. HeLa cells were cultured as described in (A), and cell cycle profiles were determined by flow cytometry at day 3. Representative flow cytometric data are depicted (left), and the bar graph and the table show quantification of flow cytometric analysis (right). Each value represents the mean ± SD of 3 independent experiments. Statistical significance is shown using the Student t test analysis; *P < 0.05; **P < 0.01; ***P < 0.001.
To confirm this, we used HeLa.S-Fucci (fluorescent ubiquitination-based cell cycle indicator) cells, which express monomeric kusabira-orange 2 (mKO2) and monomeric azami-green 1 (mAG1) fused to the ubiquitination domains of Cdt1 and geminin, respectively,Citation16 to monitor cell cycle progression in situ. As Cdt1 and geminin are the direct substrates of SCFSkp2 and APC/CCdh1 complexes, respectively, the level of Cdt1 is highest at the G1 phase, whereas geminin is prominent during S, G2, and M phases.Citation17 Therefore, the cell nuclei of HeLa.S-Fucci cells during the cell cycle are labeled with orange of mKO2 fused to the ubiquitination domain of Cdt1 in G1 phase and green of mAG1 fused to the ubiquitination domain of geminin in S, G2, and M phases. In HeLa.S-Fucci cells, siRNA-mediated knockdown of Cyclin I was confirmed by immunoblot analysis (). Two days after transfection of siRNAs for Cyclin I, we observed HeLa.S-Fucci cells with a fluorescent microscope (). Total cell number of Cyclin I siRNAs-transfected cells seemed to decrease as compared with control siRNA-transfected cells, consistent with the result of . Cells transfected with siRNAs were counted and classified into each cell cycle phase according to the criteria as shown in . Cells depleted of Cyclin I were significantly accumulated at S/G2/M phases accompanying the decrease of G1 phase (), confirming the result of flow cytometric analysis as shown in . These data strongly support the possibility that Cyclin I is involved in the cell cycle regulation at S and G2/M phases.
Figure 5. Knockdown of Cyclin I inhibits cell cycle progression in HeLa.S-Fucci cells. (A) Knockdown of Cyclin I in HeLa.S-Fucci cells. HeLa.S-Fucci cells were transfected with siRNAs for negative control (Control) or Cyclin I (Si_4, and Si_5) at day 0 and day 1, and then the expression level of Cyclin I was determined by immunoblot analysis at day 3. The protein level relative to the α-tubulin level was quantified using NIH ImageJ software. (B) Fluorescence microscopy images of Cyclin I-depleted HeLa.S-Fucci cells. HeLa.S-Fucci cells were cultured as described in (A), and then observed with fluorescence microscope at day 3. The morphology of cells and the color of cell nuclei were shown by phase contrast, mKO2, mAG1, and merged images, respectively. Bars, 50 μm. (C) The criteria for the classification of HeLa.S-Fucci cells into each cell cycle phase. Cells with mKO2 were at G1 phase; cells with both mKO2 and mAG1 were at the border of G1 and S phases; cells with mAG1 were at S, G2, and M phases; and cells with neither mKO2 nor mAG1 were at the transition from M to G1 phases, as described previously.Citation28 (D) The ratio of cell cycle phase of Cyclin I-depleted cells. HeLa.S-Fucci cells were cultured as described in (A). Cells were counted and classified into each cell cycle phase according to the criteria as shown in (C). Each value represents the mean ± SD of four counts of at least 120 cells. Statistical significance is shown using the Student t test analysis; *P < 0.05; **P < 0.01.
Discussion
In contrast to other cyclins, the Cyclin I mRNA level does not fluctuate during cell cycle progression,Citation4-Citation6 and therefore it has been believed that Cyclin I has no role in the regulation of cell proliferation. However, we showed that Cyclin I expression was regulated during the cell cycle at the protein level, and that depletion of Cyclin I prevented cell proliferation through the cell cycle arrest at S and G2/M phases.
Although Cyclin I contains the C-terminal PEST-rich regionCitation4 known as the destabilization domain,Citation13 given that PEST motif-deleted Cyclin I was still ubiquitinated (), Cyclin I is degraded by the ubiquitin-proteasome pathway, presumably mediated through the region(s) other than the PEST-rich region. Cyclin I shows the highest sequence similarity to Cyclins G1 and G2, and these two cyclins are also subjected to ubiquitination and degradation.Citation18,Citation19 Furthermore, the cyclin box of Cyclin G1 plays a role in its degradation, suggesting that the functional interaction with Cdk can regulate the stability of Cyclin G1. Therefore, it is possible that ubiquitination of Cyclin I is regulated by its interaction with Cdks or other proteins. The elevation of Cyclin I protein in several tumors was reported.Citation9,Citation10,Citation20 As the ubiquitin–proteasome pathway is essential for cell cycle regulation and cell proliferation, and dysregulation of this system leads to abnormal cell proliferation and tumor formation,Citation15 the upregulated Cyclin I protein in tumor cells may be caused by the deregulated ubiquitin-mediated protein degradation system.
It has been suggested that Cdk5, the possible binding partner of Cyclin I, plays the role in many neuronal functions, such as differentiation, migration, and cell death.Citation7,Citation8 As the activity of Cdk5 was mainly detected in postmitotic neurons, it has been considered that Cdk5 is not involved in cell cycle regulation. Recently, the relationship between Cdk5 and cell cycle has been suggested by several studies.Citation21-Citation23 Futatsugi et al. have reported that nuclear translocated Cdk5 phosphorylates Rb to release E2F transcription factor, thereby inducing cell cycle re-entry of postmitotic neurons, leading to apoptotic cell death,Citation21 whereas Zhang et al. have shown that nuclear Cdk5 associates with E2F1 to inhibit dimer formation with DP1, the E2F1 cofactor, and suppresses cell cycle entry in neurons.Citation22,Citation23 We observed that both Cyclin I and Cdk5 were detected throughout the cells (). Cytoplasmic Cdk5 in proliferating cells may have different role from nuclear Cdk5. Although the invovlement of Cdk5 in cell proliferation has not been suggested, it is possible that Cyclin I contributes to cell cycle regulation through activation of Cdk5. However, this is still an open question, because we have not detected the association of endogenous Cyclin I with Cdk5 yet. Further studies are needed to evaluate the contribution of Cyclin I to cell proliferation mediated through Cdk5.
The reciprocal functions of Cyclin I have been suggested. One is the contribution of Cyclin I to cell survival in post-differentiated and quiescent cells by activating Cdk5.Citation6 The other one is the possible involvement of Cyclin I in proliferation of cancer cells.Citation9,Citation10 The increased level of Cyclin I protein was observed in breast, ovarian, and pancreatic cancer cells.Citation9,Citation10,Citation20 Although the contribution of Cyclin I to cancer formation associated with angiogenesis was suggested,Citation9,Citation10 the precise mechanisms were unknown. In the present study, we showed the direct contribution of Cyclin I to cell proliferation by using siRNAs (). Furthermore, cells depleted of Cyclin I were accumulated at S and G2/M phases ( and ). As cell cycle checkpoint operates to determine mitotic entry by monitoring DNA replication,Citation24,Citation25 the results showing the upregulation of Cyclin I during S phase () may suggest the involvement of Cyclin I in the regulation of DNA replication. However, since the detailed molecular mechanisms by which Cyclin I regulates cell proliferation are still unknown, further investigation will be needed to clarify the role of Cyclin I in the regulation of the cell cycle.
Materials and Methods
Antibodies
Anti-Cyclin I antibody was generated by immunization of rabbits with synthetic peptide CVFRLHPSSVPGPDFSKD, corresponding to the sequence of amino acids 276–292, in which the N-terminal cysteine residue was added to allow the conjugation of the peptide to the carrier protein upon immunization. The 42 kDa band consistent with the predicted molecular weight of Cyclin I was detected on immunoblot analysis with this antibody. Anti-Flag monoclonal antibody (F3165, 1:2500), anti-Flag polyclonal antibody (F7425, 1:1000), and anti-α-tubulin monoclonal antibody (T6199, 1:1000) were obtained from SIGMA ALDRICH; Horseradish peroxidase (HRP)-conjugated anti-rabbit antibody (W4011, 1:2500 to 1:10 000) and HRP-conjugated anti-mouse antibody (W4021, 1:2500) were from Promega; anti-Cdk5 polyclonal antibody (sc-173, 1:200) and HRP-conjugated anti-rat antibody (sc-2032, 1:2500) were from Santa Cruz; anti-DsRed polyclonal antibody (632496, 1:1000) was from Clontech; anti-β-actin monoclonal antibody (#3700, 1:1000) was from Cell Signaling; anti-HA monoclonal antibodies (1666606, 1:2500 in immunoblot; 1:100 in immunoprecipitation and 1867423, 1:1000 in immunoblot; 1:125 in immunofluorescence) were from Roche; anti-Cyclin E monoclonal antibody (551160, 1:1000) was from BD Biosciences; anti-Cyclin B1 monoclonal antibody (K0128-3, 1:20 000) was from Medical and Biological Laboratories.
Plasmid constructions
The Cyclin I cDNA fragment corresponding to the amino acid sequence of full-length human Cyclin I (NCBI accession number NP_006826.1) was amplified from a human thymus cDNA library with the primers, 5′-CGCTCGAGCCACCATGAAGTTTCCAGGGCCT-3′ and 5′-CGCTCGAGCTACATGACAGAAACAGG-3′. The resultant fragment was cloned into the Xho I site of pBluescript SK(−) (Stratagene) and sequenced and then cloned into the Xho I site of pDsRed-Monomer-C1 (BD Biosciences) to construct pDsRed-Cyclin I. For construction of Flag-tagged Cyclin I expression plasmid, the Cyclin I cDNA was amplified with a pair of primers (a forward primer: 5′-CGCTCGAGATCTAAGTTTCCAGGGCCTT-3′ and a reverse primer: 5′-CGCTCGAGATCTACATGACAGAAACAGG-3′) using Cyclin I cDNA cloned in pBluescript SK(−) as a template and cloned into the Bgl II site of a pTB701-Flag to generate pTB701-Flag-Cyclin I. The fragment of PEST motif-deleted Cyclin I (Cyclin I-ΔPEST; amino acids 1–279) was amplified with the same forward primer and a reverse primer 5′-CGTGCTCGAGATCTATAATCTGAACACTCCT-3′. HA-tagged ubiquitin expression plasmid, pCGN-HA-ubiquitin, was kindly provided by Dr M Nakao (Kumamoto University). Construction of HA-tagged Cdk5 expression plasmid, pcDNA3-HA-Cdk5, was described previously.Citation26
Cell culture, transfection, and synchronization
HeLa (a cervical carcinoma cell line) cells were a gift from Dr H Sakamoto (Kobe University). HEK293T (a human embryonic kidney cell line) cells were purchased from Thermo Scientific Open Biosystems. HeLa.S-FucciCitation16 cells were obtained from Riken Cell Bank. HeLa, HEK293T, and HeLa.S-Fucci cells were cultured in DMEM supplemented with 10% fetal bovine serum. Transfection was performed using FuGENE HD (Promega) or Lipofectamine (Invitrogen) according to the manufacturer’s instructions. Lactacystin was purchased from Calbiochem and used at the final concentration of 10 μg/ml. For synchronization at late G1 phase, HeLa cells were seeded at a density of 3 × 105 cells per 60 mm dish and cultured for 24 h. After exposure to 2 mM thymidine for 18 h, cells were washed with phosphate-buffered saline (PBS) and incubated in fresh medium for 10 h and then exposed to 2 mM thymidine again for 12 h. To release cells from the late G1 arrest, cells were washed with PBS, and incubated in fresh medium for different times.
Immunoblot analysis
Cells were lysed in lysis buffer (1% Nonidet P-40, 50 mM Tris-HCl [pH 7.5], 5 mM EDTA, 150 mM NaCl, 20 mM NaF, 10 μg/ml leupeptin, 10 μg/ml aprotinin, 10 μg/ml phenylmethanesulfonyl fluoride, 10 μM MG132). The lysates and the immunoprecipitates were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and blotted onto Immobilon polyvinylidene difluoride membrane (Millipore). Each protein was detected using primary antibodies, HRP-conjugated secondary antibodies, and the ECL detection reagent (GE Healthcare).
Immunoprecipitation
For immunoprecipitation experiments, cells were lysed in lysis buffer supplemented with 1mM dithiothreitol, which prevents oxidation-induced nonspecific protein precipitation during immunoprecipitation. Lysates were incubated with 5% (vol/vol) anti-Flag M2 agarose beads (SIGMA ALDRICH) or anti-HA monoclonal antibody together with protein G Sepharose (GE Healthcare) for 2 h at 4 °C with constant rotation. The bound proteins were eluted by addition of 250 μg/ml Flag peptide or 1 mg/ml HA peptide-containing lysis buffer. Cell lysates and the eluates were separated by SDS-PAGE and immunoblotted with the indicated antibodies. For analysis of the binding of Cyclin I to Cdk5, cells were extracted by sonication in lysis buffer without Nonidet P-40, because it has been reported that Cdk5-p39 complex is dissociated by the use of non-ionic detergent.Citation27
Immunofluorescence
For immunofluorescence analysis, cells were fixed with 4% formaldehyde in PBS, permeabilized in 0.5% TritonX-100, and then incubated with primary antibodies in Can Get Signal immunostain Solution B (TOYOBO) overnight at 4 °C and incubated with Alexa Fluor 488- or Alexa Fluor 546-conjugated secondary antibodies (Molecular Probes, 1:800) for 1 h at room temperature. After staining nuclei with Hoechst 33258, cells were examined under fluorescence microscope (model BZ-8000; Keyence).
RNA interference
siRNAs for Cyclin I (Hs_CCNI_1 SI00339969, Hs_CCNI_3 SI00339983, Hs_CCNI_4 SI00339990 and Hs_CCNI_5 SI03246334) and control siRNA were obtained from Qiagen. For efficient gene silencing, siRNAs were transfected twice. HeLa or HeLa.S-Fucci cells were seeded and transfected with 30 nM siRNAs using siPORT NeoFX Transfection Agent (Applied Biosystems) according to the manufacturer’s instruction. After incubation for 24 h, second transfection was performed using HiPerFect Transfection Reagent (Qiagen) according to the manufacturer’s instruction.
Cell proliferation assay
Cell proliferation reagent WST-1 (Roche) was used as the index of cell proliferation according to the manufacture’s instruction. In brief, HeLa cells were plated at 2 × 103 cells per well in a 96-well plate and incubated for different days, followed by the addition of WST-1 reagent to medium and additional incubation for 1 h. The cleavage of the tetrazolium salt WST-1 by mitochondrial dehydrogenases was measured by absorbance at 450 nm with a reference wavelength of 650 nm.
Flow cytometry
HeLa cells were incubated with 0.5 ml of PI/RNase buffer (BD Biosciences) after fixation with 70% ethanol at −30 °C, and the DNA content was measured using FACSCalibur (Becton-Dickinson). One × 104 events were analyzed for each sample and the data were plotted using CellQuest software (Becton-Dickinson).
| Abbreviations: | ||
| Cdk | = | cyclin-dependent kinase |
| HA | = | hemagglutinin |
| siRNA | = | small interfering RNA |
| Fucci | = | fluorescent ubiquitination-based cell cycle indicator |
| mKO2 | = | monomeric kusabira-orange 2 |
| mAG1 | = | monomeric azami-green 1 |
| PBS | = | phosphate-buffered saline |
| SDS-PAGE | = | sodium dodecyl sulfate-polyacrylamide gel electrophoresis |
| HRP | = | horseradish peroxidase |
Acknowledgments
We thank Drs Hiroshi Sakamoto and Atsushi Miyawaki for HeLa and HeLa.S-Fucci cells, respectively. This work was supported in part by Grant-in-Aid for Scientific Research on Priority Areas of the Ministry of Education, Culture, Sports, Science, and Technology of Japan, and by Grant-in-Aid for Scientific Research (B) of Japan Society for the Promotion of Science.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- Murray AW. Recycling the cell cycle: cyclins revisited. Cell 2004; 116:221 - 34; http://dx.doi.org/10.1016/S0092-8674(03)01080-8; PMID: 14744433
- Morgan DO. Cyclin-dependent kinases: engines, clocks, and microprocessors. Annu Rev Cell Dev Biol 1997; 13:261 - 91; http://dx.doi.org/10.1146/annurev.cellbio.13.1.261; PMID: 9442875
- Pines J. Cyclins and cyclin-dependent kinases: theme and variations. Adv Cancer Res 1995; 66:181 - 212; http://dx.doi.org/10.1016/S0065-230X(08)60254-7; PMID: 7793314
- Nakamura T, Sanokawa R, Sasaki YF, Ayusawa D, Oishi M, Mori N. Cyclin I: a new cyclin encoded by a gene isolated from human brain. Exp Cell Res 1995; 221:534 - 42; http://dx.doi.org/10.1006/excr.1995.1406; PMID: 7493655
- Jensen MR, Audolfsson T, Factor VM, Thorgeirsson SS. In vivo expression and genomic organization of the mouse cyclin I gene (Ccni). Gene 2000; 256:59 - 67; http://dx.doi.org/10.1016/S0378-1119(00)00361-9; PMID: 11054536
- Griffin SV, Olivier JP, Pippin JW, Roberts JM, Shankland SJ. Cyclin I protects podocytes from apoptosis. J Biol Chem 2006; 281:28048 - 57; http://dx.doi.org/10.1074/jbc.M513336200; PMID: 16847066
- Hisanaga S, Endo R. Regulation and role of cyclin-dependent kinase activity in neuronal survival and death. J Neurochem 2010; 115:1309 - 21; http://dx.doi.org/10.1111/j.1471-4159.2010.07050.x; PMID: 21044075
- Kesavapany S, Li BS, Amin N, Zheng YL, Grant P, Pant HC. Neuronal cyclin-dependent kinase 5: role in nervous system function and its specific inhibition by the Cdk5 inhibitory peptide. Biochim Biophys Acta 2004; 1697:143 - 53; http://dx.doi.org/10.1016/j.bbapap.2003.11.020; PMID: 15023357
- Landberg G, Nilsson K, Jirström K, Rydén L, Kitching R, Burger AM, et al. Cyclin I is expressed in human breast cancer and closely associated with VEGF and KDR expression. Breast Cancer Res Treat 2005; 89:313 - 6; http://dx.doi.org/10.1007/s10549-004-2230-y; PMID: 15754132
- Cybulski M, Jarosz B, Nowakowski A, Jeleniewicz W, Seroczyński P, Mazurek-Kociubowska M. Cyclin I correlates with VEGFR-2 and cell proliferation in human epithelial ovarian cancer. Gynecol Oncol 2012; 127:217 - 22; http://dx.doi.org/10.1016/j.ygyno.2012.06.038; PMID: 22772062
- Liu Y, Tang MK, Cai DQ, Li M, Wong WM, Chow PH, et al. Cyclin I and p53 are differentially expressed during the terminal differentiation of the postnatal mouse heart. Proteomics 2007; 7:23 - 32; http://dx.doi.org/10.1002/pmic.200600456; PMID: 17154274
- Brinkkoetter PT, Olivier P, Wu JS, Henderson S, Krofft RD, Pippin JW, et al. Cyclin I activates Cdk5 and regulates expression of Bcl-2 and Bcl-XL in postmitotic mouse cells. J Clin Invest 2009; 119:3089 - 101; http://dx.doi.org/10.1172/JCI37978; PMID: 19729834
- Rechsteiner M, Rogers SW. PEST sequences and regulation by proteolysis. Trends Biochem Sci 1996; 21:267 - 71; PMID: 8755249
- Hershko A, Ciechanover A. The ubiquitin system. Annu Rev Biochem 1998; 67:425 - 79; http://dx.doi.org/10.1146/annurev.biochem.67.1.425; PMID: 9759494
- Nakayama KI, Nakayama K. Ubiquitin ligases: cell-cycle control and cancer. Nat Rev Cancer 2006; 6:369 - 81; http://dx.doi.org/10.1038/nrc1881; PMID: 16633365
- Sakaue-Sawano A, Kurokawa H, Morimura T, Hanyu A, Hama H, Osawa H, et al. Visualizing spatiotemporal dynamics of multicellular cell-cycle progression. Cell 2008; 132:487 - 98; http://dx.doi.org/10.1016/j.cell.2007.12.033; PMID: 18267078
- Nishitani H, Lygerou Z, Nishimoto T. Proteolysis of DNA replication licensing factor Cdt1 in S-phase is performed independently of geminin through its N-terminal region. J Biol Chem 2004; 279:30807 - 16; http://dx.doi.org/10.1074/jbc.M312644200; PMID: 15138268
- Piscopo DM, Hinds PW. A role for the cyclin box in the ubiquitin-mediated degradation of cyclin G1. Cancer Res 2008; 68:5581 - 90; http://dx.doi.org/10.1158/0008-5472.CAN-07-6346; PMID: 18632610
- Xu G, Bernaudo S, Fu G, Lee DY, Yang BB, Peng C. Cyclin G2 is degraded through the ubiquitin-proteasome pathway and mediates the antiproliferative effect of activin receptor-like kinase 7. Mol Biol Cell 2008; 19:4968 - 79; http://dx.doi.org/10.1091/mbc.E08-03-0259; PMID: 18784254
- Sun ZL, Zhu Y, Wang FQ, Chen R, Peng T, Fan ZN, et al. Serum proteomic-based analysis of pancreatic carcinoma for the identification of potential cancer biomarkers. Biochim Biophys Acta 2007; 1774:764 - 71; http://dx.doi.org/10.1016/j.bbapap.2007.04.001; PMID: 17507299
- Futatsugi A, Utreras E, Rudrabhatla P, Jaffe H, Pant HC, Kulkarni AB. Cyclin-dependent kinase 5 regulates E2F transcription factor through phosphorylation of Rb protein in neurons. Cell Cycle 2012; 11:1603 - 10; http://dx.doi.org/10.4161/cc.20009; PMID: 22456337
- Zhang J, Cicero SA, Wang L, Romito-Digiacomo RR, Yang Y, Herrup K. Nuclear localization of Cdk5 is a key determinant in the postmitotic state of neurons. Proc Natl Acad Sci USA 2008; 105:8772 - 7; http://dx.doi.org/10.1073/pnas.0711355105; PMID: 18550843
- Zhang J, Li H, Yabut O, Fitzpatrick H, D’Arcangelo G, Herrup K. Cdk5 suppresses the neuronal cell cycle by disrupting the E2F1-DP1 complex. J Neurosci 2010; 30:5219 - 28; http://dx.doi.org/10.1523/JNEUROSCI.5628-09.2010; PMID: 20392944
- Osborn AJ, Elledge SJ, Zou L. Checking on the fork: the DNA-replication stress-response pathway. Trends Cell Biol 2002; 12:509 - 16; http://dx.doi.org/10.1016/S0962-8924(02)02380-2; PMID: 12446112
- Kastan MB, Bartek J. Cell-cycle checkpoints and cancer. Nature 2004; 432:316 - 23; http://dx.doi.org/10.1038/nature03097; PMID: 15549093
- Asada A, Yamamoto N, Gohda M, Saito T, Hayashi N, Hisanaga S. Myristoylation of p39 and p35 is a determinant of cytoplasmic or nuclear localization of active cyclin-dependent kinase 5 complexes. J Neurochem 2008; 106:1325 - 36; http://dx.doi.org/10.1111/j.1471-4159.2008.05500.x; PMID: 18507738
- Yamada M, Saito T, Sato Y, Kawai Y, Sekigawa A, Hamazumi Y, et al. Cdk5--p39 is a labile complex with the similar substrate specificity to Cdk5--p35. J Neurochem 2007; 102:1477 - 87; http://dx.doi.org/10.1111/j.1471-4159.2007.04505.x; PMID: 17394551
- Hashimoto T, Juso K, Nakano M, Nagano T, Kambayashi S, Nakashima A, et al. Preferential Fas-mediated apoptotic execution at G1 phase: the resistance of mitotic cells to the cell death. Cell Death Dis 2012; 3:e313; http://dx.doi.org/10.1038/cddis.2012.52; PMID: 22622132