Abstract
Different types of mature B-cell lymphocytes are overall highly similar. Nevertheless, some B cells proliferate intensively, while others rarely do. Here, we demonstrate that a simple binary classification of gene expression in proliferating vs. resting B cells can identify, with remarkable selectivity, global in vivo regulators of the mammalian cell cycle, many of which are also post-translationally regulated by the APC/C E3 ligase. Consequently, we discover a novel regulatory network between the APC/C and the E2F transcription factors and discuss its potential impact on the G1–S transition of the cell cycle. In addition, by focusing on genes whose expression inversely correlates with proliferation, we demonstrate the inherent ability of our approach to also identify in vivo regulators of cell differentiation, cell survival, and other antiproliferative processes. Relying on data sets of wt, non-transgenic animals, our approach can be applied to other cell lineages and human data sets.
Introduction
Systematic studies of cell cycle proteins have always been a matter of great interest in basic and biomedical research, particularly due to the strong link between the mammalian cell cycle and cancer. Genome-wide approaches, typically performed in vitro or in transformed or cancer cells,Citation1,Citation2 have been instrumental in cell cycle research. Nevertheless, such methods greatly rely on drug- or serum-induced cell cycle synchronization, which inevitably introduces unwanted biochemical perturbations and, more importantly, cannot account for the full complexity of the cell cycle process in the context of multicellular organisms.
Profiling gene expression in resting and proliferative tissues can potentially reveal genes that regulate the cell cycle in vivo. Attempts to do so typically involve comparing cancerous to quiescent tissues or regenerative to post-mitotic tissues. To this end, transgenic animals expressing fluorescently-tagged cell cycle proteins have been successfully utilized to monitor proliferating cells for various in vivo applications, including genome-wide studies.Citation3,Citation4 Yet, results from such experimental models should be interpreted with caution due to the potential cytotoxicity of fluorescent proteins (especially when constitutively overexpressed) and the variables introduced by ectopically expressed tagged-cell cycle proteins.Citation5-Citation9
Analyzing 1 set of proliferating vs. resting cells may not be selective enough for identifying ubiquitous regulators of cell proliferation, because the ability to cycle is unlikely to be the only cellular property differentiating 2 cell types. Analyzing multiple proliferating cell types (e.g., regenerative skin and liver cells) vs. multiple resting cell types (e.g., post-mitotic nerve and muscle cells) can naturally overcome this limitation. On the other hand, the high degree of dissimilarity between different somatic cell types inevitably rarifies data analysis in a way that can potentially distort results and data interpretation.
To overcome these limitations, we used cell lineages as a natural source of proliferating and resting cells for the purpose of elucidating the transcriptome signature of cell proliferation in vivo. B lymphocytes play a key part in the humoral adaptive immune response in vertebrates. Despite an overall similarity, subtypes of mature B lymphocytes differ markedly in their proliferation capacity; while certain types of B cells cycle at some of the highest rates known for adult mammalian cells, others are long-lived and rarely, if ever, divide.Citation10 This feature of the B-cell lineage enables categorization of cell types on the basis of proliferation state alone, irrespective of their unique identity, exact function, and in vivo compartmentalization. Consequently, a simple binary classification of gene expression (“on” or “off”) in relation to their proliferation state can be used on B-cell expression data sets to effectively reveal global molecular circuitries regulating the cell cycle and proliferation in vivo in an unbiased manner. In principle, the exact same approach can also reveal genes that promote antiproliferative processes.
In this first report, we demonstrate the power of this conceptually simple methodology to identify both cell cycle regulators and genes controlling cell differentiation, cell survival, and other antiproliferative processes, with relevance to cancer and other human diseases.
Results
Gene expression analysis of the B-cell lineage identifies in vivo cell cycle regulators
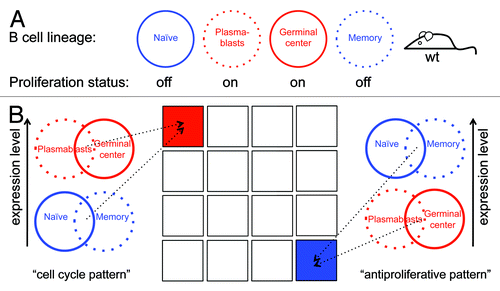
The primary aim of this study was to identify new in vivo regulators of the mammalian cell cycle using gene expression data sets originating from wt, non-transgenic mice as an unbiased source. To this end, we chose the mature B-cell lineage as a model system and used the microarray data set GSE4142,Citation11 which contains gene-expression profiles of splenic naïve, plasmablast, germinal center (GC), and memory B cells from wt mice (). We hypothesized that genes highly expressed in plasmablast and GC cells (the 2 proliferating B-cell types), but minimally expressed in naïve and memory cells (the 2 resting B-cell types), are enriched with cell cycle activators/regulators (see Supplementary Material for more details). Defining a high or low gene expression mode in 4 types of B cells will result in 16 (24) possible expression patterns, only 1 of which denotes genes whose expression is high in both types of dividing cells and low in both types of resting cells (). From here on, we refer to this expression pattern as the “cell cycle pattern”.
Figure 1. Using the B-cell lineage for identifying in vivo cell cycle regulators. (A) Splenic plasmablast and germinal center B cells are highly proliferative (red), whereas naïve and memory B-cells hardly, if ever, divide (blue). (B) Genes whose expression is switched on in both plasmablast and germinal center B cells, and switched off in both naïve and memory B cells, are likely to regulate the mammalian cell cycle and to promote proliferation in vivo. Genes with the reverse expression pattern are likely to promote antiproliferative processes in vivo. Overall, there are 16 combinatorial patterns of gene expression (illustrated as squares), of which 1 is the cell cycle pattern (red) and 1 is the antiproliferative pattern (blue).
First, we examined whether the expression profile of canonical cell cycle genes supports our hypothesis. Expression of the cyclins B1, B2, and E2 was indeed high in the proliferating cell group (plasmablast and GC cells) and low in the resting cell group (naïve and memory cells) but remarkably uniform within each group, in support of our hypothesis (). We next defined 2 phenotype classes in the microarray data, i.e., dividing and resting, and searched for the genes that correspond to our cell cycle pattern. Employing Gene Set Enrichment Analysis (GSEA) on this microarray data yielded a significant enrichment of cell cycle genes, as defined by Whitfield et al.Citation1 in their classic survey of oscillating mRNA transcripts in HeLa cells, within the subset of genes whose expression was found to be the most closely correlated with B-cell proliferation (see colored bar in ) (ES = 0.66 with nominal P value = 0 after 1000 permutations of the phenotype class labels).
Figure 2. Defining the transcriptome signature of cell proliferation. (A) Normalized expression levels (log2-intensity, arbitrary units [AU]) of the cyclins B1 (CCNB1), B2 (CCNB2), and E2 (CCNE2) in the 4 B-cell types (see ), according to GEO data set GSE4142. (B) The complete GSE4142 data set was analyzed to identify genes most closely correlated with the cell cycle pattern (see colored bar). We complemented the analysis by GSEA with default settings, indicating significant enrichment (nominal P value < 0.001) of the cell cycle gene set, as defined by Whitfield et al.Citation1 (C) A heat map showing differentially expressed genes in proliferating (plasmablast and GC) vs. resting (naïve and memory) B cells. We used the ComparativeMarkerSelection module (GenePattern) to identify differentially expressed genes based on SNR with co-variance >0.4 and FDR Q value <0.02 in the comparison of proliferating vs. resting phenotypes. Eighty-three genes are depicted as cell cycle genes, of which 30 are known APC/C targets (marked with black triangles). (D) Top 10 GO term categories (ordered by P value; all P values <0.000 01) over-represented in our list of 83 cell cycle genes (AmiGO search tool; default parameters).
![Figure 2. Defining the transcriptome signature of cell proliferation. (A) Normalized expression levels (log2-intensity, arbitrary units [AU]) of the cyclins B1 (CCNB1), B2 (CCNB2), and E2 (CCNE2) in the 4 B-cell types (see Fig. 1A), according to GEO data set GSE4142. (B) The complete GSE4142 data set was analyzed to identify genes most closely correlated with the cell cycle pattern (see colored bar). We complemented the analysis by GSEA with default settings, indicating significant enrichment (nominal P value < 0.001) of the cell cycle gene set, as defined by Whitfield et al.Citation1 (C) A heat map showing differentially expressed genes in proliferating (plasmablast and GC) vs. resting (naïve and memory) B cells. We used the ComparativeMarkerSelection module (GenePattern) to identify differentially expressed genes based on SNR with co-variance >0.4 and FDR Q value <0.02 in the comparison of proliferating vs. resting phenotypes. Eighty-three genes are depicted as cell cycle genes, of which 30 are known APC/C targets (marked with black triangles). (D) Top 10 GO term categories (ordered by P value; all P values <0.000 01) over-represented in our list of 83 cell cycle genes (AmiGO search tool; default parameters).](/cms/asset/9d45e314-fb09-4759-b685-ed680240de03/kccy_a_10926030_f0002.gif)
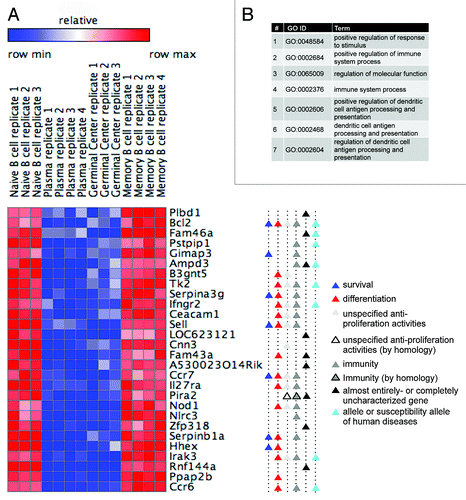
We then focused our attention on genes with the largest expression differences between the 2 phenotype classes (dividing and resting) and with the most uniform expression levels within each class. Using the ComparativeMarkerSelection module (GenePattern), we identified 171 genes, differentially expressed in proliferating vs. resting cells (Fig. S1), and annotated the best scoring 83 genes, for which we could clearly recognize the cell cycle pattern of expression as cell cycle genes ().
The top 10 gene ontology (GO) term categories most significantly over-represented in this list of cell cycle genes correspond to cell cycle-related processes only (). An in-depth inspection confirms that the majority of the 83 genes our approach identified as cell cycle genes indeed encode for proteins with well-established roles in the cell cycle, including: (1) cyclins, Cdk1, Cdc25, and UbcH10; (2) components of the E2F family of transcription factors; (3) pre-replication complex assembly proteins (e.g., Cdc6, Mcm10); (4) kinesin and kinesin-related proteins (e.g., Kif4, Kif2c); and (5) cytokinesis regulators (e.g., Aurora A and B). Importantly, during the course of this study, the 3 uncharacterized proteins 2810417H13Rik (updated name: Paf15), 4930547N16Rik (updated name: Pari), and Gas2l3 were shown to be canonical cell cycle regulators,Citation12-Citation14 emphasizing the specificity of our approach in predicting novel in vivo cell cycle regulators. Many of the 83 genes were implicated in cancer regulation, prognosis, or therapy (for example, see refs. Citation15–Citation17), illustrating the potential of our approach to also identify genes with hitherto uncharacterized cancer-related activities. Finally, only 4 (FADS1, SCN11A, GPM6A, and IgJ) out of the 83 putative cell cycle proteins for which protein function is known to some extent have yet to be linked to cell cycle regulation. These 4 genes can be theoretically interpreted as false-positive hits. We, however, suggest that these genes may hold an unappreciated role in cell cycle regulation.
Many of the genes annotated here as cell cycle genes are also known to be regulated at the post-translational level. Specifically, we noticed that 36% of our 83 cell cycle genes genes are regulated by a specific ubiquitin ligase, that is, the anaphase-promoting complex/cyclosome (APC/C), which times the degradation of canonical cell cycle proteins, thus regulating orderly cell cycle progression (, genes marked with black triangles; see references in Table S1). These APC/C targets includes the mitotic cyclins A2, B1, and B2, whose degradation, mediated by the APC/C cofactor Cdc20, regulates mitotic exit, and Aurora B and Cdc6, whose degradation, mediated by the APC/C cofactor Cdh1, regulates orderly cytokinesis and S-phase entry.Citation18-Citation20 This level of enrichment is estimated to be nearly 100-fold of expected, considering there are ~100 known APC/C targets out of 25 000 genes in the human genome. The proteins Ttk (Mps1), Cenpf, Paf15, Stil, and Gas2l3 were reported as APC/C targets during the course of this study (see supporting references in Table S1), further emphasizing the selectivity of our approach for APC/C substrates, even though it was not specifically designed for this purpose.
Transcriptome signature of cell proliferation reveals in vivo regulators of cell survival, differentiation, and other antiproliferative processes
As demonstrated in , genes whose expression is switched on in plasmablasts and GC cells and switched off in naïve and memory B cells indeed regulate the cell cycle and promote proliferation. At the other end of the spectrum are genes bearing the reverse expression pattern: switched off in plasmablasts and GC cells, and switched on in naïve and memory B cells (). By the same token, these genes are likely to promote antiproliferative cellular activities, i.e., activities that are mutually exclusive with respect to cell cycle and cell proliferation, e.g., cellular differentiation. To test our hypothesis, we looked into the filtered list of 171 differentially expressed genes (Fig. S1), and selected the 28 genes at its bottom (i.e., most dissimilar from the expression pattern shared by the cell cycle genes), for which we clearly recognized an expression pattern that inversely correlates with cell proliferation.
Unlike cell cycle genes that regulate a well-defined and distinctive process, genes regulating antiproliferative processes (denoted here as "antiproliferation genes") are expected to be: (1) more tissue-specific; (2) relatively less characterized; and (3) unrelated to cell cycle activities. GO term analysis, as well as careful survey of the literature, indeed confirmed that none of the 28 genes have been linked to proliferation-promoting activities (). Moreover, 9 genes on this list (32%) are completely uncharacterized (labeled with black triangles in ). Nearly all the other 19 genes are established components of the immune system (labeled with dark gray triangles), of which 7 are confirmed alleles or susceptibility alleles of human diseases, as inferred by the OMIM database (labeled with cyan triangles). Importantly, 15 of the 19 characterized genes, such as the homeobox gene Hhex, have at least one established link to cell differentiation (see supporting references in Table S2), primarily, albeit not exclusively, in the context of B or immune cells.Citation21 An additional gene, Fam43a, whose function is unknown, has been reported to be upregulated during keratinocyte differentiation.Citation22 Interestingly, 7 of the 19 characterized genes (37%) positively regulate cell survival (labeled with blue triangles), not necessarily in the context of B or immune cells. Most notable is the canonical cell survival factor Bcl2.Citation23,Citation24 In addition, other antiproliferative activities, not necessarily related to differentiation or survival (, gray triangles), have been reported for more than half of the 19 characterized genes, including Bcl-2 and Ifngr2, reducing proliferation; Cnn3 (Calponin 3), cell-cell fusion; and Pira2, neuronal plasticity (see Table S2 for supporting references). Altogether, we demonstrate that profiling gene expression in proliferating vs. resting B cells can reveal, in addition to cell cycle genes, also in vivo regulators of antiproliferative processes, particularly of cell differentiation and cell survival.
Figure 3. Profiling gene expression in proliferating vs. resting B cells reveals in vivo regulators of cell differentiation and survival. (A) A heat map showing the expression of 28 differentially expressed genes in proliferating and resting B cells (for more details, see ; Fig. S1). Gene functions are depicted (see Table S2 for references). (B) The top 7 GO term categories (ordered by P value) over-represented in the genes listed in (A) (AmiGO search tool; default parameters; all P values are below 0.008 5).
A novel regulatory link between APC/CCdh1 and the atypical branch of the E2F family of transcription factors
E2F1, the canonical member of the “activating” branch of the E2F family of transcription factors, is profoundly linked to cell proliferation by triggering the transcription of a large group of S- and M-phase genes. In contrast, E2F7 and E2F8, which make up the atypical, repressive branch of the E2F family, function in post-mitotic cells and regulate cell differentiation and apoptosis.Citation25,Citation26 Nevertheless, the E2F7 and E2F8 transcripts oscillate throughout the cell cycle, as they are both direct targets of E2F1. Moreover, both genes repress E2F1 expression and activity, forming a negative feedback loop.Citation25,Citation27,Citation28 As expected, E2F1 expression in the GSE4142 data set was indeed correlative to cell proliferation: high in the proliferating GC and plasmablast B cells, and low in the resting naïve and memory B cells (). However, to our surprise, out of the E2F members recorded in the GSE4142 data set, only E2F7 and E2F8 met the stringent criteria initially set for selecting cell cycle genes (, no. 4 and no. 43, respectively), showing differential expression that matches that of cyclins ( and ).
Figure 4. APC/CCdh1 targets E2F7 and E2F8 for degradation. (A) Normalized expression levels of E2F1, E2F7, E2F8, and cyclin A2 (CCNA2) in the depicted B-cell types, according to GEO database GSE4142. (B) The proteins XlTome-1, hE2F7, and hE2F8 were expressed in rabbit reticulocytes supplemented with 35S-methionine, and incubated in G1 phase S3 cell extracts supplemented with wt or dominant negative (DN) Ubch10 and either buffer, MG132, the C-terminus of Emi1, or unlabeled Securin. Time-dependent degradation was assayed by SDS-PAGE and autoradiography (top), and quantified (bottom). (C) HEK293 cells were co-transfected with Cdh1 or empty vector (pCS2) and with either E2F8, or E2F7-EGFP, or EGFP, at a 4:1 ratio. After 30 h, cells were harvested for western blotting with E2F8, GFP (experimental control), and tubulin (loading control) antibodies. (B and C) are representative experiments. (D and E) Model for the interplay between APC/CCdh1 and E2F1, E2F7, and E2F8. APC/CCdh1 regulates the proteolysis of both E2F1 and its repressors, E2F7 and E2F8 (B and C) during G1-phase. The model proposes that a delicate difference in the timing of proteolysis of the activating vs. repressive E2F transcription factors controls the G1/S transition of the mammalian cell cycle (see main text for further information/discussion).
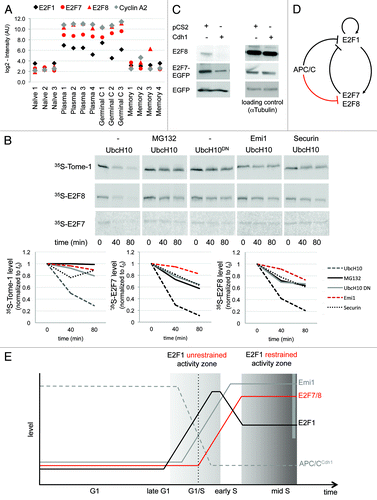
In light of the high level of enrichment of APC/C targets in our list of cell cycle genes () and the fact that E2F1 itself,Citation29 as well as many of its targets, is degraded during the G1-phase by APC/CCdh1, we next tested whether E2F7 and E2F8 are also regulated by this complex. To this end, E2F7 and E2F8, as well as the 2 established APC/CCdh1 substrates Tome-1 and Securin,Citation30,Citation31 were in vitro translated (IVT) in the presence of 35S-methionine. The IVT products were then incubated in active human-cell extracts prepared from G1-synchronous HeLa S3 cells. This cell-free system is widely regarded as suitable for identifying APC/C substrates.Citation2 Both E2F7 and E2F8, as well as Tome-1 and Securin (being used here as positive controls), were effectively degraded in G1 extracts supplemented with the UbcH10, the E2 partner of APC/C, but not in the presence of: (1) the proteasome inhibitor MG132; (2) UbcH10DN, a dominant-negative mutant of UbcH10; (3) the APC/C specific inhibitor Emi1; and (4) an excess of unlabeled Securin, being used here as a specific competitive inhibitor of APC/CCdh1 (; Fig. S2). In addition, the overexpression of Cdh1 in HEK293T cells lowered the levels of both E2F7 and E2F8 proteins (). Altogether, these findings demonstrate that E2F7 and E2F8 are novel substrates of APC/CCdh1.
Discussion
Having the same E3 complex to downregulate E2F1 and its 2 repressors, E2F7 and E2F8, seems counterintuitive (). However, considering the time dimension of the cell cycle, these results, in fact, could account for the rise in E2F1 levels and activity in late G1, despite the negative feedback imposed by its own targets, E2F7 and E2F8 (). In the late G1-phase, E2F1 accumulates in the nucleus, bound to its dimerization partner (DP) protein,Citation32 which protects it from APC/CCdh1-mediated degradation, thus enabling its accumulation even in the presence of the active complex.Citation33 We show that E2F7 and E2F8 are also APC/CCdh1 substrates (). However, both proteins lack the DP binding domains, are not protected by DP proteinsCitation25 and thus can be maintained at a low level as long as APC/CCdh1 is active. By activating its own transcription,Citation34,Citation35 E2F1 levels and activity can now rapidly increase, triggering the expression of its targets Emi1 and Cyclin A2,Citation36,Citation37 which blockCitation38 and disassembleCitation39 the APC/CCdh1 complex, a hallmark of the G1/S transition. Only then, following APC/C inactivation, E2F7 and E2F8 can accumulate and negatively regulate E2F1, restraining its activity throughout the S- and G2-phases. Hence, the unrestrained activity of E2F1 is required for pushing cells into the S-phase; however, once cells traverse G1/S, E2F1 activity is balanced by E2F7 and E2F8, or otherwise cells might undergo apoptosis ().Citation40,Citation41
Our strategy for profiling the cell cycle transcriptome is based on the natural humoral immune response and is conceptually simple. By classifying gene expression with respect to proliferation state (), our approach yielded a selective list of cell cycle genes enriched with APC/C targets and cancer-promoting genes (). An important feature of our approach is its inherent ability to also identify genes whose expression inversely correlates with cell proliferation ( and ). These genes either repress proliferation directly or promote cellular processes that are essentially mutually exclusive with cell proliferation. In practice, our approach has proven highly selective for identifying positive regulators of cell differentiation as well as cell survival. While the former is not compatible with proliferation in many ways, the latter reinforces the notion that pro-survival activities are essential for prolonging the lifespan of long-lived, resting cells, such as naïve and memory B cells, though are dispensable in cycling cells with a typical life cycle of 10 to 24 h. Interestingly, a genome-scale inverse correlation between cell proliferation and survival has been reported in bacteria.Citation42 There is no simple way to estimate the proportion of genes regulating cell differentiation and survival in the human/mouse genome. Moreover, many of the genes we have discovered in this category are entirely uncharacterized. These hurdles emphasize the potential of our new approach to reveal novel molecular circuitries that regulate cell differentiation, cell survival, and additional antiproliferation processes, beyond the context of immunity.
Our approach has several major advantages: (1) it is in vivo and non-perturbative; (2) it can be applied to human models as well as to other cell lineages; (3) it does not require chemically or mechanically-induced cellular synchronizations; (4) it does not rely on transgenic- or mutant animals; (5) nor does it depend on potentially cytotoxic and immunogenic fluorescent protein tags.Citation5,Citation43 Moreover, the use of 2 sets of highly proliferative and resting B cells is particularly important: first, the overall similarity between mature B cells simplifies any genome-wide analysis. Second, the 4 cell types enable 16 binary expression patterns, of which only 1 represents cell cycle genes and only 1 represents antiproliferation genes. Using this ratio of 1 to 16 gains our methodology a selectivity that is 4-fold higher than had we used only 1 set of proliferating vs. resting cells. Furthermore, our approach truly enables the filtering out of differentially expressed genes that are unrelated to either pro- or antiproliferation activities per se. Reducing the overall rate of false-positive results is particularly advantageous in the context of cell differentiation and other antiproliferation activities, of which our understanding is limited compared with that of the cell cycle.
Our approach is demonstrated here using a platform data set of protein-coding genes. Applying our analysis to comprehensive deep-sequencing data sets (M Vecsler, M Cohen, and A Tzur, unpublished data) can reveal entirely new pools of transcripts that either regulate the mammalian cell cycle in vivo, with potential roles in cancer biology, or, alternatively, promote antiproliferation activities, with clear relevance to cell differentiation and cell survival.
Materials and Methods
Microarray data analysis
The Affymetrix microarray data set GSE4142Citation11 was normalized by gcRMA in R BioConductor.Citation44 Throughout this study, we analyzed this data set by comparing expression profiles from proliferating (plasmablasts and GC) vs. resting (naive and memory) B cells. We used ComparativeMarkerSelection module from GenePatternCitation45 to detect significantly differentially expressed genes according to the following criteria: covariance >0.4 of their expression values within each phenotype and signal to noise ratios (SNR) with FDR Q values <0.02.Citation46 Functional enrichment analysis of the differentially expressed genes in the space of gene ontology (GO) terms was performed using the AmiGO search tool with default parameters. We applied Gene Set Enrichment Analysis (GSEA)Citation47 to this data set in order to test the enrichment of a set of cell cycle genes derived from a study of synchronously dividing HeLa cells.Citation1
Plasmids
Open reading frames (ORF) of human E2F7 and E2F8 were amplified from U2OS (human osteosarcoma cells) cDNA using primers flanked by FseI (forward) and AscI (reverse) sites. The ORFs of E2F8 and E2F7 were cloned into pCS2-FA vector for in vitro (SP6 promoter) and in vivo (CMV promoter) expression. For cloning into the EGFP-N1 vector (Clontech Laboratories, Inc), ORF of E2F7 was amplified with primers flanked by HindIII (forward) and AgeI (reverse). The expression cassette of the E27-EGFP was cloned into the pCDNA4/TO (Invitrogen) using HindIII and NotI for transfection. The expression vectors: pGEX-4T-3-Emi1 C terminus (amino acids 299–447), pET28-UbcH10, pET28-UbcH10C114S (UbcH10DN), pET28-hSecurin, pCS2-hSecurin, pCS2-Tome-1, pCS2-Cdh1, pCS2-Cdc20, and pCS2-FA were a gift from Dr Marc Kirschner (Harvard Medical School) and described elsewhere. We used the pEGFP-N1 vector (Clontech Laboratories, Inc) to express EGFP in cells. All transfections were performed using the Metafectene transfection reagent (Biontex Laboratories GmbH) according to the manufacturer’s protocol.
Tissue culture and cell synchronization
HEK293T and HeLa S3 (S3) cells were maintained in DMEM (Gibco) supplemented with 10% fetal bovine serum (Gibco) and penicillin and streptomycin (Gibco) at 37 °C, 5% CO2. For G1 extract preparation, S3 cells were grown in suspension (1L spinner flask, 85 rpm) until population reached a density of approximately 5 × 105 cells/ml. The cell culture was then incubated with 2 mM thymidine (Sigma-Aldrich) for 20–22 h, washed, and released into fresh media for 3–3.5 h, and then blocked again with 50 ng/ml nocodazole (Sigma-Aldrich) for 12 h. Cells were then washed twice with fresh, warm media, released for 3 h, and harvested to generate G1 (APC/CCdh1-active) extracts.
Cell extracts preparation
G1 cell extracts were prepared as follows: synchronized S3 cells were harvested, washed with PBS, lysed in swelling buffer (20 mM HEPES, pH 7.7, 2 mM MgCl2, 5 mM KCl, 1 mM DTT, and EDTA-free protease inhibitor cocktail), supplemented with energy-regeneration mix (1 mM ATP, 7.5 mM creatine phosphate, 70 μg/ml creatine phosphokinase, and 0.1 mM EGTA), and homogenized by freeze-thawing and passage through 20 G needle. Extracts were cleared by subsequent centrifugations (5 min at 2700 × g; 45 min at 18 000 × g) and stored at −80 °C. Extracts for western blotting analysis were prepared following a standard protocol.
Degradation assay
Degradation assays were performed using 20 μl G1 cell extracts supplemented with 1 μl of 20 × energy-regeneration mix, 1 μl of 10 mg/ml Ubiquitin (Boston Biochem), 1 μl of 10 mg/ml recombinant UbcH10 or UbcH10DN, 5 μL of either 1 mg/ml Emi1 C-terminus fragment, 3.5 mg/ml Securin, or PBS, and 1–2 μl radiolabeled IVT product expressed in reticulocytes (Promega) with 35S-labeled methionine (PerkinElmer). Samples were incubated at 30 °C. Aliquots were taken at 0, 40, and 80 min, removed into sample buffer, and snap-frozen on dry ice. Samples were resolved by SDS-PAGE, visualized by autoradiography, and quantified (ImageQuant).
Antibodies
The following antibodies were used: anti-α-Tubulin (Thermo Fisher: MS581P1), anti-E2F8 (Abnova: H00079733-M01), and anti-GFP (Santa Cruz: sc-9996).
Additional material
Download Zip (483.4 KB)Acknowledgments
We are deeply indebted to Dr Jane Seagal for introducing us to the wonders of immunology, and to Prof Doron Ginsberg for reagents. Funding by the Israeli Centers of Research Excellence (I-CORE), Gene Regulation in Complex Human Disease, Center No. 41/11 (AT); the Israel Cancer Association Grant 20120067 (AT); the German–Israeli Foundation (GIF), No. 2294-2269.2/2011 (AT); and the Israel Cancer Research Fund (ICRF) postdoctoral fellowship (MV) is gratefully acknowledged.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Supplementary Materials
Supplementary materials may be found here: www.landesbioscience.com/journals/cc/article/26030
References
- Whitfield ML, Sherlock G, Saldanha AJ, Murray JI, Ball CA, Alexander KE, Matese JC, Perou CM, Hurt MM, Brown PO, et al. Identification of genes periodically expressed in the human cell cycle and their expression in tumors. Mol Biol Cell 2002; 13:1977 - 2000; http://dx.doi.org/10.1091/mbc.02-02-0030.; PMID: 12058064
- Ayad NG, Rankin S, Ooi D, Rape M, Kirschner MW. Identification of ubiquitin ligase substrates by in vitro expression cloning. Methods Enzymol 2005; 399:404 - 14; http://dx.doi.org/10.1016/S0076-6879(05)99028-9; PMID: 16338372
- Klochendler A, Weinberg-Corem N, Moran M, Swisa A, Pochet N, Savova V, Vikeså J, Van de Peer Y, Brandeis M, Regev A, et al. A transgenic mouse marking live replicating cells reveals in vivo transcriptional program of proliferation. Dev Cell 2012; 23:681 - 90; http://dx.doi.org/10.1016/j.devcel.2012.08.009; PMID: 23000141
- Sakaue-Sawano A, Kurokawa H, Morimura T, Hanyu A, Hama H, Osawa H, Kashiwagi S, Fukami K, Miyata T, Miyoshi H, et al. Visualizing spatiotemporal dynamics of multicellular cell-cycle progression. Cell 2008; 132:487 - 98; http://dx.doi.org/10.1016/j.cell.2007.12.033; PMID: 18267078
- Liu HS, Jan MS, Chou CK, Chen PH, Ke NJ. Is green fluorescent protein toxic to the living cells?. Biochem Biophys Res Commun 1999; 260:712 - 7; http://dx.doi.org/10.1006/bbrc.1999.0954; PMID: 10403831
- Taghizadeh RR, Sherley JL. CFP and YFP, but not GFP, provide stable fluorescent marking of rat hepatic adult stem cells. J Biomed Biotechnol 2008; 2008:453590; http://dx.doi.org/10.1155/2008/453590; PMID: 18401450
- Baens M, Noels H, Broeckx V, Hagens S, Fevery S, Billiau AD, Vankelecom H, Marynen P. The dark side of EGFP: defective polyubiquitination. PLoS One 2006; 1:e54; http://dx.doi.org/10.1371/journal.pone.0000054; PMID: 17183684
- Goto H, Yang B, Petersen D, Pepper KA, Alfaro PA, Kohn DB, Reynolds CP. Transduction of green fluorescent protein increased oxidative stress and enhanced sensitivity to cytotoxic drugs in neuroblastoma cell lines. Mol Cancer Ther 2003; 2:911 - 7; PMID: 14555710
- Link CD, Fonte V, Hiester B, Yerg J, Ferguson J, Csontos S, Silverman MA, Stein GH. Conversion of green fluorescent protein into a toxic, aggregation-prone protein by C-terminal addition of a short peptide. J Biol Chem 2006; 281:1808 - 16; http://dx.doi.org/10.1074/jbc.M505581200; PMID: 16239215
- Janeway C, Travers P, Walport M, Shlomchik M. Immunobiology. Gerald Science, 2004.
- Luckey CJ, Bhattacharya D, Goldrath AW, Weissman IL, Benoist C, Mathis D. Memory T and memory B cells share a transcriptional program of self-renewal with long-term hematopoietic stem cells. Proc Natl Acad Sci U S A 2006; 103:3304 - 9; http://dx.doi.org/10.1073/pnas.0511137103; PMID: 16492737
- Emanuele MJ, Ciccia A, Elia AE, Elledge SJ. Proliferating cell nuclear antigen (PCNA)-associated KIAA0101/PAF15 protein is a cell cycle-regulated anaphase-promoting complex/cyclosome substrate. Proc Natl Acad Sci U S A 2011; 108:9845 - 50; http://dx.doi.org/10.1073/pnas.1106136108; PMID: 21628590
- Moldovan GL, Dejsuphong D, Petalcorin MI, Hofmann K, Takeda S, Boulton SJ, D’Andrea AD. Inhibition of homologous recombination by the PCNA-interacting protein PARI. Mol Cell 2012; 45:75 - 86; http://dx.doi.org/10.1016/j.molcel.2011.11.010; PMID: 22153967
- Wolter P, Schmitt K, Fackler M, Kremling H, Probst L, Hauser S, Gruss OJ, Gaubatz S. GAS2L3, a target gene of the DREAM complex, is required for proper cytokinesis and genomic stability. J Cell Sci 2012; 125:2393 - 406; http://dx.doi.org/10.1242/jcs.097253; PMID: 22344256
- Görgün G, Calabrese E, Hideshima T, Ecsedy J, Perrone G, Mani M, Ikeda H, Bianchi G, Hu Y, Cirstea D, et al. A novel Aurora-A kinase inhibitor MLN8237 induces cytotoxicity and cell-cycle arrest in multiple myeloma. Blood 2010; 115:5202 - 13; http://dx.doi.org/10.1182/blood-2009-12-259523; PMID: 20382844
- Ray D, Kiyokawa H. CDC25A phosphatase: a rate-limiting oncogene that determines genomic stability. Cancer Res 2008; 68:1251 - 3; http://dx.doi.org/10.1158/0008-5472.CAN-07-5983; PMID: 18316586
- Takezawa K, Okamoto I, Tsukioka S, Uchida J, Kiniwa M, Fukuoka M, Nakagawa K. Identification of thymidylate synthase as a potential therapeutic target for lung cancer. Br J Cancer 2010; 103:354 - 61; http://dx.doi.org/10.1038/sj.bjc.6605793; PMID: 20628382
- Mailand N, Diffley JF. CDKs promote DNA replication origin licensing in human cells by protecting Cdc6 from APC/C-dependent proteolysis. Cell 2005; 122:915 - 26; http://dx.doi.org/10.1016/j.cell.2005.08.013; PMID: 16153703
- Lindon C, Pines J. Ordered proteolysis in anaphase inactivates Plk1 to contribute to proper mitotic exit in human cells. J Cell Biol 2004; 164:233 - 41; http://dx.doi.org/10.1083/jcb.200309035; PMID: 14734534
- Peters JM. The anaphase promoting complex/cyclosome: a machine designed to destroy. Nat Rev Mol Cell Biol 2006; 7:644 - 56; http://dx.doi.org/10.1038/nrm1988; PMID: 16896351
- Hunter MP, Wilson CM, Jiang X, Cong R, Vasavada H, Kaestner KH, Bogue CW. The homeobox gene Hhex is essential for proper hepatoblast differentiation and bile duct morphogenesis. Dev Biol 2007; 308:355 - 67; http://dx.doi.org/10.1016/j.ydbio.2007.05.028; PMID: 17580084
- Paragh G. Novel insights into transcriptional changes during keratinocyte differentiation and ultraviolet radiation response. Doctoral Thesis, 2009, Semmelweis University, Budapest, Hungury.
- McDonnell TJ, Deane N, Platt FM, Nunez G, Jaeger U, McKearn JP, Korsmeyer SJ. bcl-2-immunoglobulin transgenic mice demonstrate extended B cell survival and follicular lymphoproliferation. Cell 1989; 57:79 - 88; http://dx.doi.org/10.1016/0092-8674(89)90174-8; PMID: 2649247
- Middleton G, Wyatt S, Ninkina N, Davies AM. Reciprocal developmental changes in the roles of Bcl-w and Bcl-x(L) in regulating sensory neuron survival. Development 2001; 128:447 - 57; PMID: 11152643
- Lammens T, Li J, Leone G, De Veylder L. Atypical E2Fs: new players in the E2F transcription factor family. Trends Cell Biol 2009; 19:111 - 8; http://dx.doi.org/10.1016/j.tcb.2009.01.002; PMID: 19201609
- DeGregori J. E2F and cell survival: context really is key. Dev Cell 2005; 9:442 - 4; http://dx.doi.org/10.1016/j.devcel.2005.09.006; PMID: 16198285
- Endo-Munoz L, Dahler A, Teakle N, Rickwood D, Hazar-Rethinam M, Abdul-Jabbar I, Sommerville S, Dickinson I, Kaur P, Paquet-Fifield S, et al. E2F7 can regulate proliferation, differentiation, and apoptotic responses in human keratinocytes: implications for cutaneous squamous cell carcinoma formation. Cancer Res 2009; 69:1800 - 8; http://dx.doi.org/10.1158/0008-5472.CAN-08-2725; PMID: 19223542
- Moon NS, Dyson N. E2F7 and E2F8 keep the E2F family in balance. Dev Cell 2008; 14:1 - 3; http://dx.doi.org/10.1016/j.devcel.2007.12.017; PMID: 18194644
- Budhavarapu VN, White ED, Mahanic CS, Chen L, Lin FT, Lin WC. Regulation of E2F1 by APC/C Cdh1 via K11 linkage-specific ubiquitin chain formation. Cell Cycle 2012; 11:2030 - 8; http://dx.doi.org/10.4161/cc.20643; PMID: 22580462
- Ayad NG, Rankin S, Murakami M, Jebanathirajah J, Gygi S, Kirschner MW. Tome-1, a trigger of mitotic entry, is degraded during G1 via the APC. Cell 2003; 113:101 - 13; http://dx.doi.org/10.1016/S0092-8674(03)00232-0; PMID: 12679038
- Zou H, McGarry TJ, Bernal T, Kirschner MW. Identification of a vertebrate sister-chromatid separation inhibitor involved in transformation and tumorigenesis. Science 1999; 285:418 - 22; http://dx.doi.org/10.1126/science.285.5426.418; PMID: 10411507
- Bandara LR, Buck VM, Zamanian M, Johnston LH, La Thangue NB. Functional synergy between DP-1 and E2F-1 in the cell cycle-regulating transcription factor DRTF1/E2F. EMBO J 1993; 12:4317 - 24; PMID: 8223441
- Peart MJ, Poyurovsky MV, Kass EM, Urist M, Verschuren EW, Summers MK, Jackson PK, Prives C. APC/C(Cdc20) targets E2F1 for degradation in prometaphase. Cell Cycle 2010; 9:3956 - 64; http://dx.doi.org/10.4161/cc.9.19.13162; PMID: 20948288
- Hsiao KM, McMahon SL, Farnham PJ. Multiple DNA elements are required for the growth regulation of the mouse E2F1 promoter. Genes Dev 1994; 8:1526 - 37; http://dx.doi.org/10.1101/gad.8.13.1526; PMID: 7958837
- Neuman E, Flemington EK, Sellers WR, Kaelin WG Jr.. Transcription of the E2F-1 gene is rendered cell cycle dependent by E2F DNA-binding sites within its promoter. Mol Cell Biol 1994; 14:6607 - 15; PMID: 7935380
- Tategu M, Nakagawa H, Sasaki K, Yamauchi R, Sekimachi S, Suita Y, Watanabe N, Yoshid K. Transcriptional regulation of human polo-like kinases and early mitotic inhibitor. J Genet Genomics 2008; 35:215 - 24; http://dx.doi.org/10.1016/S1673-8527(08)60030-2; PMID: 18439978
- Timmers C, Sharma N, Opavsky R, Maiti B, Wu L, Wu J, Orringer D, Trikha P, Saavedra HI, Leone G. E2f1, E2f2, and E2f3 control E2F target expression and cellular proliferation via a p53-dependent negative feedback loop. Mol Cell Biol 2007; 27:65 - 78; http://dx.doi.org/10.1128/MCB.02147-05; PMID: 17167174
- Reimann JD, Gardner BE, Margottin-Goguet F, Jackson PK. Emi1 regulates the anaphase-promoting complex by a different mechanism than Mad2 proteins. Genes Dev 2001; 15:3278 - 85; http://dx.doi.org/10.1101/gad.945701; PMID: 11751633
- Sørensen CS, Lukas C, Kramer ER, Peters JM, Bartek J, Lukas J. A conserved cyclin-binding domain determines functional interplay between anaphase-promoting complex-Cdh1 and cyclin A-Cdk2 during cell cycle progression. Mol Cell Biol 2001; 21:3692 - 703; http://dx.doi.org/10.1128/MCB.21.11.3692-3703.2001; PMID: 11340163
- Zalmas LP, Zhao X, Graham AL, Fisher R, Reilly C, Coutts AS, La Thangue NB. DNA-damage response control of E2F7 and E2F8. EMBO Rep 2008; 9:252 - 9; http://dx.doi.org/10.1038/sj.embor.7401158; PMID: 18202719
- Li J, Ran C, Li E, Gordon F, Comstock G, Siddiqui H, Cleghorn W, Chen HZ, Kornacker K, Liu CG, et al. Synergistic function of E2F7 and E2F8 is essential for cell survival and embryonic development. Dev Cell 2008; 14:62 - 75; http://dx.doi.org/10.1016/j.devcel.2007.10.017; PMID: 18194653
- Shoval O, Sheftel H, Shinar G, Hart Y, Ramote O, Mayo A, Dekel E, Kavanagh K, Alon U. Evolutionary trade-offs, Pareto optimality, and the geometry of phenotype space. Science 2012; 336:1157 - 60; http://dx.doi.org/10.1126/science.1217405; PMID: 22539553
- Gambotto A, Dworacki G, Cicinnati V, Kenniston T, Steitz J, Tüting T, Robbins PD, DeLeo AB. Immunogenicity of enhanced green fluorescent protein (EGFP) in BALB/c mice: identification of an H2-Kd-restricted CTL epitope. Gene Ther 2000; 7:2036 - 40; http://dx.doi.org/10.1038/sj.gt.3301335; PMID: 11175316
- Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol 2004; 5:R80; http://dx.doi.org/10.1186/gb-2004-5-10-r80; PMID: 15461798
- Kuehn H, Liberzon A, Reich M, Mesirov JP. Using GenePattern for gene expression analysis. Curr Protoc Bioinformatics 2008; Chapter 7:Unit 7 12.
- Golub TR, Slonim DK, Tamayo P, Huard C, Gaasenbeek M, Mesirov JP, Coller H, Loh ML, Downing JR, Caligiuri MA, et al. Molecular classification of cancer: class discovery and class prediction by gene expression monitoring. Science 1999; 286:531 - 7; http://dx.doi.org/10.1126/science.286.5439.531; PMID: 10521349
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 2005; 102:15545 - 50; http://dx.doi.org/10.1073/pnas.0506580102; PMID: 16199517