Abstract
Interactions between RNA binding proteins (RBPs) and genes are not well understood, especially in regulation of angiogenesis. The RBP HuR binds to the AU-rich (ARE) regions of labile mRNAs, facilitating their translation into protein and has been hypothesized to be a tumor-maintenance gene. Elevated levels of cytoplasmic HuR directly correlate with increased invasiveness and poor prognosis for many cancers, including those of the breast. HuR controls the expression of multiple genes involved in angiogenesis including VEGFa, HIF1a, and thrombospondin 1 (TSP1). We investigated the role of HuR in estrogen receptor negative (ER-) breast cancer. MDA-MB-231 cells with higher levels of HuR have alterations in cell cycle kinetics and faster growth. Unexpectedly, HuR overexpression significantly interfered with tumor growth in orthotopic mouse models. The putative mechanism seems to be an anti-angiogenetic effect by increasing expression of TSP1 but also surprisingly, down-regulation of VEGF, a target of HuR which it normally increases. Our findings reveal that HuR may be regulating a cluster of genes involved in blood vessel formation which controls tumor angiogenesis. An approach of modulating HuR levels may overcome limitations associated with monotherapies targeting tumor vessel formation.
Introduction
Although most breast cancers are estrogen receptor positive (ER+), approximately 15% are estrogen receptor negative (ER−).Citation1 In general, ER− tumors are more aggressive, have a poor clinical outcome and dismal survival rates that disproportionately affect lower income and minority women. The poor prognosis is partly attributed to the fact that these women are limited to conventional cytotoxic chemotherapy. This is in distinct contrast to ER+ tumors, which can be treated with hormonal therapies such as tamoxifen or aromatase inhibitors. Moreover, the presence of the growth factor HER2 allows oncologists to employ targeted therapy using HER2 receptor-specific drugs.
The role of posttranscriptional gene regulation in cancer is now widely appreciated, as the contributions of RNA binding proteins (RBPs) and microRNAs (miRNAs) are becoming more apparent.Citation2–Citation8 The posttranscriptional operon hypothesis states that RBPs and miRNAs are coordinately regulating expression of biologically related mRNAs. This has generated interest in identification of additional gene products in the acquired capabilities model of malignant transformation.Citation9 The RBP HuR, a paraneoplastic antigen, overexpressed in many malignancies including breast cancer, has been implicated as an important RBP which may function as a tumor maintenance gene, facilitating malignant transformation.Citation10–Citation17 HuR has been shown to regulate genes in multiple areas of the acquired capabilities model, including two pivotal genes involved in angiogenesis, VEGF and HIF1α.Citation18–Citation22 Increased HuR cytoplasmic expression directly correlates with severity and aggressiveness of many cancers, including those of the breast.Citation23–Citation25 HuR has been demonstrated to be an important prognostic factor in familial breast cancer patients.Citation23,Citation24 Recently, investigators altered the subcellular distribution of HuR and documented a decrease in mRNA stability, subsequent ER levels and susceptibility to tamoxifen.Citation26
In this study we investigated the role of HuR in ER− breast cancer by gain-of-function ER− (MDA-MB-231) cell lines which overexpressed epitope tagged hemagglutinin (HA) HuR. As expected and consistent with published results for other cancer types, HuR overexpression in vitro resulted in increasing MDA-MB-231 cellular growth rates and alterations in cell cycle kinetics. Surprisingly, when these cells were used in vivo as xenografts in athymic mice, a 90% reduction in growth rate was seen, as compared to control groups. The presumptive mechanism was an anti-angiogenic affect by increasing the expression of thrombospondin 1 (TSP1), a gene known to be anti-angiogenic. However, unexpectedly, there were also decreases in VEGF expression. Thus, our results indicate that HuR overexpression in ER− breast cancer can potently inhibit tumor growth by potentially blocking angiogenesis.
Results
Overexpression of HuR in MDA-MB-231 cells increases growth rates and alters cell cycle kinetics.
To study the role of HuR expression in MDA-MB-231 ER− breast cancer we made individual clones which overexpressed either epitope tagged (HA) HuR or empty vector (EV) control and measured growth rates and cell cycle kinetics (). Overexpression of HA-HuR resulted in accelerated cellular growth as determined by counting (). When the cells were stained with propidium iodide, we noted an alteration in cell cycle kinetics (). HA-HuR overexpressing cells had increased amounts of cells in G1 (67 vs. 57%), as compared with empty vector controls. Conversely, HuR overexpression also resulted in a compensatory decrease in G2/M percentages (18% vs. 27%). We concluded, as expected, that HuR overexpression resulted in increases in growth rates of MDA-MB-231 cells. We then investigated the effects of HuR overexpression in vivo, using orthotopic xenograft animal models.
HuR overexpression results in significantly reduced tumor growth and mass.
The clones used in , empty vector (2C7) and overexpresser HA-HuR (4E1), were injected into the contralateral mammary fat pads of athymic nude mice. Tumor growth was assessed weekly by caliper measurements and followed in vivo by MRI scan. Surprisingly, tumors overexpressing HuR had significant inhibition of growth, whereas EV control tumors increased significantly in tumor volume (). The tumors were removed from the animals on day 42. Histological staining, western blotting and RT-PCR were performed to determine whether any cells remained. As seen in Supplementary Data, both control (2C7) and HuR (4E1) tumors had intact human GAPDH mRNA and tumor 4E1 expressed the HA-HuR transgene. Furthermore, the HA-HuR tumors harvested on day 42 still expressed HA-HuR protein (Fig. S1). We reestablished cell lines from tumors removed from animals to study their growth rates. We found that these reestablished cells had similar growth rates to parental cells prior to transplantation (Fig. S2).
The experiments were repeated with wild-type, parental MDA-MB-231 cells to validate these findings. As seen in , both parental and control cells (2C7) grew at similar rates, whereas tumors which overexpressed HuR (4E1) did not appear to grow (). Tumor mass was assessed and showed that HuR overexpression resulted in a 90% reduction in growth (). These results were confirmed by MRI scans, gross photographs and microscopy ( and D). HA-HuR tumors appeared to be a gelatin-like capsule and the control tumors were a solid round mass. Cross sections of both revealed that the HA-HuR tumors had a smooth, homogeneous and glistening surface, whereas the control tumors had a heterogeneous, yellow-white surface with a necrotic center (). Both tumors contained viable cancer cells and similar morphology determined to be moderately to poorly differentiated carcinoma, consistent with the implanted MDA-MB-231 cells (). We concluded that HuR overexpression resulted in significantly smaller ER− tumors in animals.
To verify that these results were not clonal, we repeated the orthotopic tumor injection experiments using a second HuR overexpressing clone (5F1) which in vitro grew similarly to the original overexpressing clone, 4E1 (). As seen in Figure S3, tumor 5F1 also exhibited retarded growth rates as compared to empty vector controls and had a 90% reduction in mass (Fig. S3). Taken together, these findings demonstrated that HuR overexpression in MDA-MB-231 cells resulted in significant reductions in tumor growth in a clonal independent fashion.
Gene ontology (GO) analysis of overexpressed genes in HuR overexpressing cells.
In order to better understand the genes which may be involved in altering tumor growth in HuR overexpressing MDA-MB-231 cells, we performed genome wide microarray analysis (Fig. S4 and ). As seen in Figure S4, many genes were over-represented (reflected in large odds ratios) that deal with both biological processes as well as molecular function. Given the large numbers of genes, we decided to investigate the following three potential mechanisms to explain the large discrepancy seen in tumor growth: (1) increased apoptosis, (2) increases in senescence and (3) alteration in angiogenesis. The microarray profiling, however, did not distinguish between direct and indirect HuR targets. Therefore, we decided to further investigate known HuR targets related to cancer.
Tumors which overexpress HuR have decreased angiogenesis.
We conducted experiments which targeted well known HuR target genes involved in angiogenesis which had previously been described in the literature: thrombospondin 1 (TSP1), VEGF and HIF1α.Citation21,Citation22,Citation27 We therefore performed real-time PCR to measure mRNA levels of TSP1, VEGF and HIF1α. As seen in , HuR overexpression caused an increase in TSP1 mRNA and protein () but surprisingly decreases in steady-state VEGF mRNA and protein levels; HIF1α steady-state mRNA levels appeared unchanged (). We further measured HIF1α protein by western and immunohistochemical staining, which were unchanged between HuR overexpressing and control tumors (data not shown). Furthermore, RNA immunoprecipitation (RIP) using an antibody specific to HuR and an IgG1 antibody control revealed that HuR interacts with TSP1 and VEGF mRNA, as these transcripts were 38- and 54-fold enriched, respectively, in the HuR IP pellet, as compared with istotype control. TSP1 and VEGF mRNA were upregulated 62- and 59-fold in the HuR IP from the HuR overexpression cells. GAPDH, which does not interact with HuR showed no enrichment in either HuR IP when compared to the IgG1 IP (). After confirming HuR interacts with TSP1 and VEGF mRNA we sought to gain a better understanding of the mechanism by which the overexpression of HuR leads to increases in TSP1 mRNA and protein and decreases in VEGF mRNA and protein. To determine whether alterations in HuR levels were altering TSP1 and VEGF mRNA stability we performed mRNA decay assays using actinomycin D. The VEGF mRNA stability was not altered when comparing HuR overexpressing cells with empty vector control (). TSP1 mRNA half-life was increased in the HuR overexpressing cells when compared to the empty vector control cells ().
We next examined whether alterations in apoptosis could account for differences in tumor growth. To determine whether HuR overexpression was altering apoptosis in vitro we performed both annexin V and 7-AAD staining. No differences in amount of cells undergoing apoptosis between the HuR overexpressers and EV controls were seen ( and S5). In vivo, there were no increases in apoptosis in HuR overexpression tumors, as compared with EV control tumors harvested on day 42 post-inoculation ( and S6). Increased apoptosis in EV tumors was mostly found in the necrotic centers. To account for the possibility that apoptosis was occurring early in tumor formation, we repeated the experiments and harvested tumors from mice 14 days post-inoculation. No increases in apoptosis were seen in HA-HuR tumors as compared to EV control tumors ( and S6). The tumors from mice 14 days post-inoculation were only beginning to be visible. Hematoxylin and eosin staining revealed tumor morphology consistent with breast adenocarcinoma (Fig. S7). We further quantitated the amount of blood vessels and found a significant decrease in vessel formation in HA-HuR tumors as compared with EV controls ( and C). No tortuous vessels were seen. These non-functional vessels are sometimes seen when there are perturbations in the DLL4-Notch signaling pathways. Measurement of senescence using β-galactosidase staining did not reveal any significant differences (Fig. S8). Taken together, we concluded that HuR overexpression in MDA-MB-231 tumors does not appear to alter cellular apoptosis but may interfere with angiogenesis by overexpression of an anti-angiogenetic factor, TSP1 and by downregulation of a known pro-angiogenic factor, VEGF.
Discussion
In ER− cells overexpressing HuR, there are increases in growth rates in vitro as well as alterations in cell cycle kinetics. Specifically, MDA-MB-231 cells overexpressing HuR have increases in the G1 phase of the cell cycle, consistent with published results.Citation25 A plausible explanation is HuR induced stabilization of cyclin B1, the pivotal cyclin involved in G2/M transition.Citation28 Surprisingly, when these same cells were transplanted into athymic nude mice, HuR overexpression resulted in a 90% growth reduction in tumor size. These results were confirmed by measuring tumor volume and mass. Additionally, the results were validated by serial MRI scans during the experimental period and further confirmed in two independent clones. Empty vector (EV) control and parental wild type MDA-MB-231 cells grew similarly and resulted in much larger tumors than those formed by HA-HuR overexpressing cells. Histological staining of cross sections from EV and HA-HuR tumors revealed them to be poorly differentiated carcinomas, consistent with ER− breast cancer. Moreover, the HuR transgene was still expressed in the smaller tumors at the time of harvest (day 42).
We searched for putative mechanisms to explain these findings, since in other systems HuR overexpression results in larger, more robust tumor growth.Citation25 No inflammatory infiltrates were seen in either tumor. As expected, there was increased apoptosis in the centers of the EV control tumors, since these regions are relatively more hypoxic. Apoptosis early in tumor development is an unlikely mechanism of inhibited tumor growth in the tumors overexpressing HuR as we did not see any alterations in apoptosis 14 days post-inoculation using two methods of apoptosis detection. Furthermore, significant apoptosis was not observed in either cell lines (in vitro) or tumors (in vivo) overexpressing HuR. These results more than likely rule out apoptosis as a mechanism of reduced tumor growth in HA-HuR tumors, although we cannot definitely exclude apoptosis earlier than 14 days. Since the tumors continue to overexpress HuR even at day 42, it would seem unlikely that HuR overexpression would be the etiology of cell death. We also ruled out senescence as a mechanism, as there was less β-galactosidase staining in the smaller tumors.
Due to previously published data on the role of HuR in controlling angiogenesis via interactions with VEGF and HIF1α mRNAs, we investigated the relationship between HuR overexpression and these pro-angiogenetic factors.Citation21,Citation22 There was a statistically significant decrease in VEGF mRNA and protein expression and no increases in HIF1α mRNA or protein expression. As expected, increased HuR expression correlated with increased TSP1 mRNA expression and protein levels. TSP1 is a well known anti-angiogenetic factor and has been described to be regulated by HuR.Citation27 Enumeration of angiogenesis by CD34 staining confirmed that HuR overexpression resulted in significant decreases in new blood vessel formation. This decrease in neovascularization may, in part, explain why tumors formed by HA-HuR overexpression were much smaller than EV controls.
Other groups have shown that decreases in VEGF expression in a variety of cancer models can inhibit angiogenesis and tumor growth.Citation29–Citation32 It has also been demonstrated that TSP1 can mediate tumor growth suppression via blocking angiogenesis, consistent with our results.Citation33,Citation34
As previously mentioned HuR has been described to stabilize TSP1 and VEGF mRNA resulting in greater levels and increased protein expression.Citation21,Citation27 In contradistinction to published reports of HuR stabilization of VEGF mRNA, our findings indicate that under conditions of HuR overexpression, there are decreases in both VEGF mRNA and protein levels. The reasons for this discrepancy are presently unknown, though the results may be dependent upon levels of HuR overexpression and hypoxia. HuR's relationship, however, with HIF1α is more complex. HuR binds to AU-rich regions in HIF1α 5′UTR, rather than the 3′UTR (even though both regions of the molecule possess AREs), and causes a translational upregulation in HIF1α protein synthesis without altering mRNA levels. It appears HuR overexpression is not affecting HIF1α protein production in our system. HIF1α is the major transcriptional factor for VEGF mRNA transcription, and as such, this is an area of intensive investigation in our laboratory.
The exact mechanisms of HuR-induced anti-angiogenetic effects are not completely understood but may involve interactions between HuR and microRNAs. In a recently published report, HuR actively recruited let-7 miRNA to c-myc mRNA, which resulted in decreases of its translation.Citation35 Furthermore, we do not have any direct evidence that the HA tag (located at the amino terminal) affects the distribution or targeting of HuR to its mRNA targets. Published results from the Kontoyonnis lab used the same epitope tag to make a transgenic mouse overexpressing HA-HuR in macrophages and although this is a different system than ours, they did not see any alterations in nuclear vs. cytoplasmic HuR distribution nor binding to its mRNA targets.Citation36 Moreover, the metastatic potential of anti-angiogenetic MDA-MB-231 HuR altered cell lines needs to be investigated, since this phenotype results in patient mortality and the correlation between tumor size and metastatic potential may not be direct. We have identified putative markers of metastasis which have been altered in vivo in these cells and this area is currently under investigation.
Interactions between RBPs such as HuR and their downstream mRNA targets are complex. Even though HuR can theoretically bind 8% of human mRNAs which contain AREs, it does not necessarily do so.Citation37–Citation39 Published reports from our lab and others demonstrate that HuR critically regulates myogenesis by controlling the expression of three central genes in this process.Citation40,Citation41 HuR siRNA knock down prevents muscle formation, whereas HuR overexpression results in precocious differentiation.Citation40,Citation41 Yet, it is highly likely HuR is interacting with many more targets inside these cells. The exact explanations for this discrete phenotype are not presently understood. Our work involving targets discovered by RIP-Chip in cell lines under normoxic conditions indicates that HuR may indeed regulate the same targets in opposite fashion, dependent upon cellular milieu, though the reasons are not presently clear.Citation42 There are obvious differences when performing these experiments under the hypoxic conditions which operate in animals in which tumors form. The published data has both indicated that HuR overexpression correlates with increased as well as decreased aggressiveness in breast cancer.Citation23,Citation24,Citation43 These results would lend credence to the idea that HuR may be directing its mRNA targets in cell-specific fashion.
In summary, our findings indicate that HuR overexpression in ER− breast cancer results in a concomitant increase in TSP1 and a decrease in VEGF expression as well as substantial decreases in tumor size. Presently, the reasons for decreases in VEGF expression in HuR-overexpressing tumors are unclear. Our results are highly reproducible when each mouse is compared with its cohorts within individual and duplicate experiments. Furthermore, it is clone-independent, in that we obtained similar results using two different overexpression clones. However, we do not yet know if these results are specific to only ER− breast cancer, or also include the ER+ subtype, as our results deal primarily with one cell line and must be validated in other cell lines before we can make generalized conclusions.
It will be interesting to test the therapeutic paradigm whereby exogenous HuR overexpression in already established tumors may abrogate their development, perhaps by interfering with angiogenesis. If correct, HuR may be involved in regulating a cluster of genes involved in angiogenesis and therefore, alterations in HuR expression may influence multiple downstream genes which operate in blood vessel formation. These experiments are currently underway. There has been a great deal of interest in blocking angiogenesis, either by increasing expression of anti-angiogenetic factors, such as TSP1 or blocking pro-angiogenetic factors such as VEGF. It has been challenging to work with TSP1 due to its large molecular size. The emerging evidence in cancer treatment approaches in this area, however, has been that monotherapies can be more readily overcome by cancer cells, which are constantly evolving in the patient. We may have discovered a novel way of limiting ER−tumor growth by interfering with neo-angiogenesis at multiple steps by modulating the expression of a key RBP involved in tumor growth.
Materials and Methods
Cell line and growth conditions.
The MDA-MB-231 cell line was purchased from American Type Culture Collection (Manassas, VA) and grown according to the vendor's directions.
Generation of clones expressing HA-HuR.
Hemagglutinin (HA) tagged human HuR was cloned into the NheI and XhoI sites of the pZeoSV2 (−) (Invitrogen™) vector. Cells were then plated and transfected with either pZeo HA-HuR or pZeo empty vector using Lipofectamine 2000 (Invitrogen™). Selection was performed with 200 µg/ml of Zeocin antibiotic (Invitrogen™) and cells were cloned by limiting dilution.
SDS-PAGE and western blot analysis.
Western analysis was performed as described previously.Citation12 For detection of VEGFα and TSP1 from tumors, triple-detergent RIPA lysis buffer with protease inhibitors was used. Membranes were probed with anti-TSP1 (Abcam) or anti-VEGFα (Abcam) and anti-β-tubulin (Sigma-Aldri+ch). Secondary antibodies used were: sheep antimouse HRP (GE Healthcare) or donkey anti-rabbit HRP (GE Healthcare) for VEGFα and TSP1, respectively. Proteins were detected using chemiluminescence (GE Healthcare). HuR, TSP1 and VEGFα levels were determined using Bio-Rad's Quantity One software (Bio-Rad) normalizing to β-tubulin. Anti-HuR 3A2 hybridoma was kindly provided by Joan Steitz (Yale School of Medicine).
In vitro growth, apoptosis and cell cycle assay.
Fifty thousand cells were seeded in a 24-well plate. Cells were then counted using a hemocytometer and trypan blue exclusion dye 24, 48 and 72 hours post-seeding n = 4 assays. For cell cycle analysis, cells were fixed and permeabilized, re-suspended in PBS with 0.2 mg/ml RNase A (Sigma-Aldrich) and 10 mg/ml propidium iodide (Sigma-Aldrich), and analyzed on FACScan (BD Biosciences, San Jose, CA) and cell cycle analysis was performed using Cell Quest software (BD Biosciences). Annexin V and 7-AAD staining was performed using the FITC Annexin V Apoptosis Detection Kit I (BD Bioscience) by following the manufacturer's protocol. Flow cytometry was performed on the FACScan (BD Biosciences) and analysis was performed using Cell Quest software (BD Biosciences).
Mice tumor inoculations and measurements.
Female athymic nude mice (aged 8–12 weeks) were purchased from Harlan. For tumor inoculations, 100 µL of a 1:1 mixture of Matrigel (BD Biosciences) and RPMI 1640 (GIBCO®) containing 1 × 106 MDA-MB-231 cells, expressing either pZeo HA-HuR, pZeo empty vector or wild-type clones, were injected into the left or right mammary fat pad. Tumor volumes were calculated using calipers by measuring using the formula: L×W×D×0.5. Procedures were conducted according to the University of Missouri Columbia Animal Care and Use Committee.
Longitudinal MRI investigation and tumor volume analysis.
Magnetic Resonance Imaging (MRI) was performed on a 7T/210 mm Varian Unity Inova MRI system (Varian Inc., Palo Alto, CA) using previously published procedures.Citation44 Mice were anesthetized with 1–2% isoflurane in oxygen via a nose cone over the entire imaging period. Three mice were imaged weekly for 5 weeks to monitor tumor growth. Mice were imaged to obtain axial planes using multi-slice spin-echo T1-weighted (T1W) imaging sequence applied with a fat-saturation pulse to suppress the strong signals from fatty tissues in the chest. Spin-echo diffusion-weighted imaging (DWI) with b-value = 1,063 s/mm2 was performed at week 4 to assess the tumor tissue viability, i.e., necrotic tissue or solid tumor tissue. Images were obtained with slice thickness = 0.8 mm and in-plane resolution = 0.059 mm × 0.156 mm. The tumors were manually segmented using VnmrJ software (Varian Inc.,) to obtain the tumor volume in cm3. DW images at week 4 were used to differentiate between necrotic tissues and solid tumor tissues.
RNA purification and real-time PCR.
RNA was extracted from snap frozen tumors in Trizol reagent (Invitrogen™). One µg of RNA was reverse transcribed and real-time PCR was performed using cDNA (in triplicate) for using SuperScript III twostep qRT-PCR with SYBR green (Invitrogen™). Primers were as follows:
VEGF sense 5′-TTT CTG CTG TCT TGG GTG CAT TGG-3′ and antisense 5′-ACC ACT TCG TGA TGA TTC TGC CCT-3′, TSP1 sense 5′-TTC CGC CGA TTC CAG ATG ATT CCT-3′ and antisense 5′-ACG AGT TCT TTA CCC TGA TGG CGT-3′, HIF1α sense 5′-TTG GCA GCA ACG ACA CAG AAA CTG-3′ and antisense 5′-TTG AGT GCA GGG TCA GCA CTA CTT-3′, GAPDH sense 5′-AGC CTC AAG ATC ATC AGC AAT GCC-3′ and antisense 5′-TGT GGT CAT GAG TCC TTC CAC GAT-3′. PCR reactions were performed using Applied Biosystems StepOne PCR system. Results were analyzed using comparative CT method with GAPDH as endogenous reference control.
Microarray.
0.5 µg of total RNA was used to make the biotin-labeled antisense RNA (aRNA) target using the Illumina TotalPrep RNA amplification kit (Ambion, Austin, TX). and hybridized to Illumina HumanWG-6 (V2_0_R3) beadchip (47,000 genes) as per Illumina protocols. After hybridization, the chips were washed and stained with streptavidin-C3 and analyzed by BeadArray reader (Illumina, San Diego, CA).
Histology and imunohistochemistry.
Tissue was routinely processed, formalin-fixed and paraffin embedded for hematoxylin-and-eosin-staining and immunohistochemistry. Immunostaining was performed using the avidin-biotin-peroxidase complex method. Slides were incubated at room temperature with the following antibodies: anti-cleaved caspase-3 antibody ([2305-PC-100], Trevigen, Gaithersburg, MD); anti-CD34 (ab8158, MEC 14.7, Abcam, Cambridge, MA); anti-HIF1α (NB100-134, Novus Biologicals, Littleton, CO); anti-VEGF (sc-152 Santa Cruz Biotechnology, Inc., Santa Cruz, CA); and anti-TSP-1 (clone A6.1, MS-420-P1, Thermo Fisher Scientific, Fremont, CA). Slides labeled with anti-CD34 or TSP-1 were incubated with a biotinylated secondary antibody DAKO for CD-34 and rabbit anti-mouse IgG (for TSP-1) followed by streptavidin-linked HRP (DAKO). Cleaved caspase-3 and VEGF slides were incubated with a HRP antibody (EnVision™, DAKO). Bound antibodies were visualized with peroxidase substrates: DAB 3, 3′-diaminobenzidine solution (DAKO) or NovaRED™ (Vector Labs, Burlingame, CA). For the TUNEL apoptosis detection assay we used the In Situ Cell Death Detection Kit, TMR red (Roche Applied Science) and followed the manufacturer's protocol for paraffin sections.
Statistics.
All error bars represent standard error of the mean. The p values were calculated using the two-tailed Student t test.
Statistical analysis of microarray data.
Analysis of microarray gene expression data was primarily performed using the Linear Models for Microarray Data (limma) packageCitation45 and the lumi package,Citation46 available through the Bioconductor projectCitation47 for use with R statistical software. Quantile normalization was used for between chip normalization. Statistical analysis was performed using moderated t-statistics applied to the log-transformed (base 2) normalized intensity for each gene. Because two measurements were taken from each mouse, the dependency between paired measurements was accounted for by a modified mixed linear model that treated each animal as a block. The within-block correlations were constrained to be equal between genes, and then information was borrowed across genes to moderate the standard deviations between genes via an empirical Bayes method. The contrast of interest computed and tested was the difference between overexpressor and control vector. Adjustment for multiple testing was made using the false discovery rate (FDR) method of Benjamini and Hochberg.Citation48 We chose 10% as our FDR-cutoff for declaring statistical significance, and a threshold of at least three-fold (up or down) for declaring a biologically significant change in expression. Gene ontology (GO) analyses were carried to test the association between Gene Ontology Consortium terms and the list of differentially expressed genes. In defining the gene universe for the analysis, non-specific filtering was used to increase statistical power without biasing the results. This filtering selected only probes on the Illumina array which had both an Entrez gene identifier,Citation49 a GO annotation (as provided in the lumiHumanAll.dbCitation50 annotation data package and GO.dbCitation51 annotation maps) and an interquartile range of ≥0.1 on log2 scale across all samples. Using this gene universe, GOstatsCitation52 was used to carry out conditional hypergeometric tests which exploit the hierarchical nature of the relationships among the GO terms for conditioning.Citation53 We carried out GO analyses for over-representation of biological process (BP), molecular function (MF) and cellular component (CC) ontologies and computed the nominal hypergeometric probability for each GO category. These results were used to assess whether the number of selected genes associated with a given term was larger than expected and a p-value cutoff of 1% was used. GO categories containing less than 10 genes from our gene universe were not considered to be reliable indicators and are not reported.
RNA immunoprecipitation.
RNA immunoprecipitation was performed as previously described.Citation42
Abbreviations
| ER− | = | estrogen receptor negative |
| ER+ | = | estrogen receptor positive |
| elav | = | embryonic abnormal vision |
| RBP | = | RNA-binding proteins |
| RIP | = | RNA immunoprecipitation |
| miRNA | = | microRNA |
Figures and Tables
Figure 1 Overexpression of HA-HuR in MDA-MB-231 cancer cells increases cell growth and alters cell cycle kinetics in vitro while inhibiting tumor growth in vivo. (A) MDA-MB-231 cells transfected with the pZeoSV2 vector expressing HA-HuR, selected with Zeocin and cloned by limiting dilution express HA-HuR compared to a pZeoSV2 empty vector control. MDA-MB-231 clones 4e1 and 5F1 expressed 42% and 38% more HuR than control as shown in representative western blot. Bar graph represents mean HuR percent overexpression from three different western blots. (B) Both clones expressing HA-HuR proliferated significantly faster than the empty vector control in vitro. (C) Overexpression of HA-HuR increased cells in G0/G1 cell cycle, from 57.70% to 67.67%. Overexpression of HA-HuR decreased cells in G2/M phase by 27.19% to 18.29% but did not significantly alter cells in S phase as measured by DNA content. (D) MDA-MB-231 HA-HuR 4E1 showed significantly reduced tumor volume (mm3) and growth starting at two weeks post-inoculation and continuing for five weeks when compared to empty vector MDA-MB-231 controls as measured by both MRI and calipers. For tumor experiments nine animals per group were used. For both counting assay and tumor volumes data represent mean value ± SEM. p < 0.05. Flow cytometry data represent mean value ± SEM from n = 4 separate experiments done in triplicate. p < 0.05.
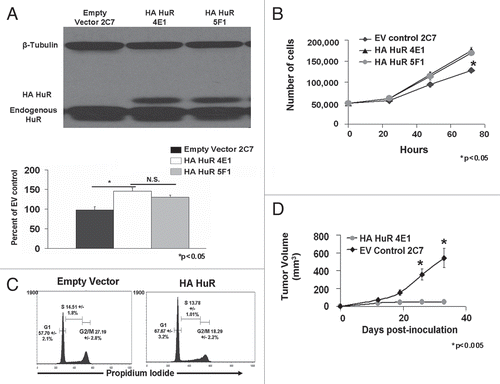
Figure 2 Overexpression of HA-HuR in MDA-MB-231 cancer cells inhibits tumor growth in athymic nude mice. (A) Repeat experiments comparing MDA-MB-231 HA-HuR 4E1 with both wild-type MDA-MB-231 and empty vector MDA-MB-231 confirmed HuR overexpression reduced tumor volume (mm3) and growth starting at two weeks post-inoculation and continuing for five weeks as measured by calipers. (B) Tumors overexpressing HA-HuR had significantly less mass after harvest 42 days post-inoculation when compared to the WT or empty vector (EV) controls. (C) MRI comparing largest sections for each tumor showed significantly smaller tumors in the HA-HuR overexpressing tumors when compared to EV control tumors. (D) Representative cross sections of tumors showed that those formed by inoculation with HA-HuR resembled a gelatin-like capsule, were significantly smaller and more homogeneous than those formed by inoculation with empty vector. Hematoxylin and eosin stain revealed poorly differentiated carcinomas with similar morphology and lack of inflammatory cells in both HA-HuR tumors and EV control tumors. Five animals were used per group in HA-HuR, empty vector and wild-type control groups. Experiments were repeated with similar results using a different clone, 5F1, (see Supporting Information Fig. S3). Data represent mean value ± SEM. p < 0.05; in photomicrographs bar = 27 microns.
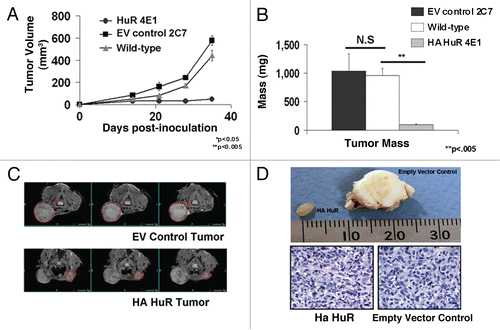
Figure 3 TSP1 is upregulated in HA-HuR tumor and VEGFα is downregulated. (A) Real-time PCR indicates TSP1 is upregulated in tumors (5.44 fold) and cells in culture (4.88 fold) overexpressing HA-HuR when compared to EV control tumors and cells, consistent with the microarray data. VEGF is downregulated (2.6 fold) in tumors overexpressing HA-HuR when compared to EV controls. HIF1α mRNA levels did not appreciably change. Change in gene expression was determined using the comparative CT method and is represented as fold change in HA-HuR tumors as compared to empty vector controls. GAPDH was used as an endogenous control. (B) Western blot for TSP1 shows increased protein expression of TSP1 (76%) in the HA-HuR overexpressing tumors when compared to EV control tumors. (C) Western blot for VEGF shows decrease protein expression by 23% in the HA-HuR overexpressing tumors when compared to EV control tumors (representative of two independent sets of tumors). Data represent mean value ± SEM from n = 3 separate mice done in triplicate. p < 0.05.
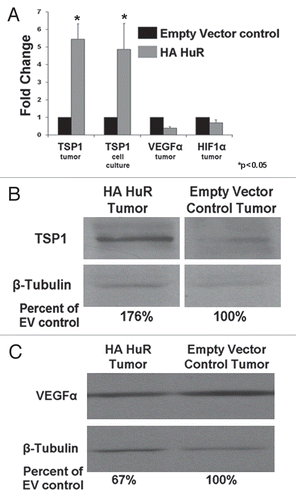
Figure 4 HuR interacts with both TSP1 and VEGF mRNAs in cells overexpressing HuR. (A) RNA immunoprecipiation indicates both TSP1 and VEGF mRNA are increased in the HuR IP when compared to IgG1 control IP in both HA-HuR overexpressing cells and EV control cells. (B) Actinomycin D mRNA stability assay shows VEGF mRNA half-life was not altered between cells overexpressing HA-HuR and EV control cells. (C) TSP1 mRNA from cells overexpressing HA-HuR has a longer half-life than TSP1 mRNA from EV control cells. For RNA immunoprecipitation, data represents mean value ± SEM. p < 0.05.
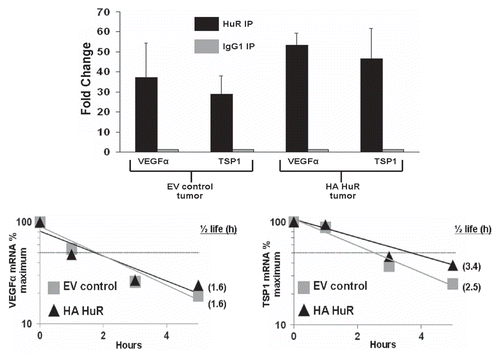
Figure 5 Tumors overexpressing HA-HuR have no increases in apoptosis but decreased blood vessel formation compared to control. (A) Annexin V staining reveals similar amounts of cells undergoing apoptosis between cells overexpressing HA-HuR and EV control cells. (B) Caspase 3 staining shows no differences in the amount of apoptosis in the EV control tumors compared to HA-HuR tumors harvested 14 days post-inoculation. In the tumors harvested on day 42 post-inoculation, caspase 3 staining showed more apoptotic cells in the EV control tumors compared to tumors overexpressing HA-HuR. CD34 staining shows fewer blood vessels in tumors overexpressing HA-HuR. (B) Quantitation of blood vessels stained (number of vessels per high power field scored) with CD34 indicated significantly fewer blood vessels in the tumors overexpressing HA-HuR. Error bars ± SEM; p < 0.005; in photomicrographs bar = 27 microns. Representative of n = 5 sets of tumors.
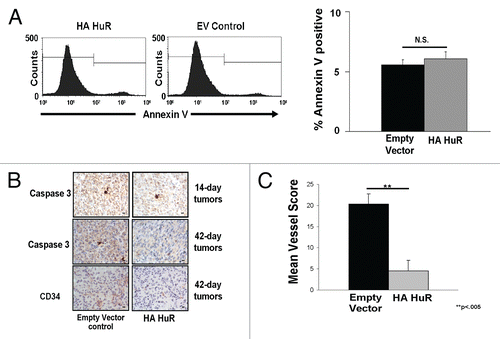
Table 1 Genes upregulated in tumors overexpressing HuR
Additional material
Download Zip (520.2 KB)Acknowledgements
We gratefully acknowledge the support provided by the VA Biomolecular Imaging Center at the Harry S. Truman VA Hospital and the University of Missouri. We would like to thank Sharon Stack and George Davis for their help in review of the manuscript. This study was supported by Department of Defense Idea Award (W81XWH-07-1-040) and University of Missouri institutional funds.
References
- Reis-Filho JS, Tutt AN. Triple negative tumours: a critical review. Histopathology 2008; 52:108 - 118
- Deng S, Calin GA, Croce CM, Coukos G, Zhang L. Mechanisms of microRNA deregulation in human cancer. Cell Cycle 2008; 7:2643 - 2646
- Esquela-Kerscher A, Slack FJ. Oncomirs—microRNAs with a role in cancer. Nat Rev Cancer 2006; 6:259 - 269
- Keene JD. Why is Hu where? Shuttling of early-response-gene messenger RNA subsets. Proc Natl Acad Sci USA 1999; 96:5 - 7
- Keene JD. Ribonucleoprotein infrastructure regulating the flow of genetic information between the genome and the proteome. Proc Natl Acad Sci USA 2001; 98:7018 - 7024
- Keene JD. Posttranscriptional generation of macromolecular complexes. Mol Cell 2003; 12:1347 - 1349
- Keene JD. Organizing mRNA export. Nat Genet 2003; 33:111 - 112
- Keene JD, Tenenbaum SA. Eukaryotic mRNPs may represent posttranscriptional operons. Mol Cell 2002; 9:1161 - 1167
- Hanahan D, Weinberg RA. The hallmarks of cancer. Cell 2000; 100:57 - 70
- Gorospe M. HuR in the mammalian genotoxic response: post-transcriptional multitasking. Cell Cycle 2003; 2:412 - 414
- Abdelmohsen K, Kim MM, Srikantan S, Mercken EM, Brennan SE, Wilson GM, et al. miR-519 suppresses tumor growth by reducing HuR levels. Cell Cycle 2010; 9
- Atasoy U, Watson J, Patel D, Keene JD. ELAV protein HuA (HuR) can redistribute between nucleus and cytoplasm and is upregulated during serum stimulation and T cell activation. J Cell Sci 1998; 111:3145 - 3156
- Dalmau J, Furneaux HM, Gralla RJ, Kris MG, Posner JB. Detection of the anti-Hu antibody in the serum of patients with small cell lung cancer—a quantitative western blot analysis. Ann Neurol 1990; 27:544 - 552
- Dalmau J, Furneaux HM, Rosenblum MK, Graus F, Posner JB. Detection of the anti-Hu antibody in specific regions of the nervous system and tumor from patients with paraneoplastic encephalomyelitis/sensory neuronopathy. Neurology 1991; 41:1757 - 1764
- Fan XC, Steitz JA. Overexpression of HuR, a nuclear-cytoplasmic shuttling protein, increases the in vivo stability of ARE-containing mRNAs. EMBO J 1998; 17:3448 - 3460
- Ma WJ, Cheng S, Campbell C, Wright A, Furneaux H. Cloning and characterization of HuR, a ubiquitously expressed Elav-like protein. J Biol Chem 1996; 271:8144 - 8151
- Lopez de Silanes I, Lal A, Gorospe M. HuR: posttranscriptional paths to malignancy. RNA Biol 2005; 2:11 - 13
- Abdelmohsen K, Lal A, Kim HH, Gorospe M. Posttranscriptional orchestration of an anti-apoptotic program by HuR. Cell Cycle 2007; 6:1288 - 1292
- Goldberg-Cohen I, Furneauxb H, Levy AP. A 40-bp RNA element that mediates stabilization of vascular endothelial growth factor mRNA by HuR. J Biol Chem 2002; 277:13635 - 13640
- Levy AP. Hypoxic regulation of VEGF mRNA stability by RNA-binding proteins. Trends Cardiovasc Med 1998; 8:246 - 250
- Levy NS, Chung S, Furneaux H, Levy AP. Hypoxic stabilization of vascular endothelial growth factor mRNA by the RNA-binding protein HuR. J Biol Chem 1998; 273:6417 - 6423
- Galban S, Kuwano Y, Pullmann R Jr, Martindale JL, Kim HH, Lal A, et al. RNA-binding proteins HuR and PTB promote the translation of hypoxia-inducible factor 1alpha. Mol Cell Biol 2008; 28:93 - 107
- Heinonen M, Bono P, Narko K, Chang SH, Lundin J, Joensuu H, et al. Cytoplasmic HuR expression is a prognostic factor in invasive ductal breast carcinoma. Cancer Res 2005; 65:2157 - 2161
- Heinonen M, Fagerholm R, Aaltonen K, Kilpivaara O, Aittomaki K, Blomqvist C, et al. Prognostic role of HuR in hereditary breast cancer. Clin Cancer Res 2007; 13:6959 - 6963
- Lopez de Silanes I, Fan J, Yang X, Zonderman AB, Potapova O, Pizer ES, et al. Role of the RNA-binding protein HuR in colon carcinogenesis. Oncogene 2003; 22:7146 - 7154
- Hostetter C, Licata LA, Witkiewicz A, Costantino CL, Yeo CJ, Brody JR, et al. Cytoplasmic accumulation of the RNA binding protein HuR is central to tamoxifen resistance in estrogen receptor positive breast cancer cells. Cancer Biol Ther 2008; 7
- Mazan-Mamczarz K, Hagner PR, Corl S, Srikantan S, Wood WH, Becker KG, et al. Post-transcriptional gene regulation by HuR promotes a more tumorigenic phenotype. Oncogene 2008; 27:6151 - 6163
- Wang W, Caldwell MC, Lin S, Furneaux H, Gorospe M. HuR regulates cyclin A and cyclin B1 mRNA stability during cell proliferation. EMBO J 2000; 19:2340 - 2350
- Raskopf E, Vogt A, Sauerbruch T, Schmitz V. siRNA targeting VEGF inhibits hepatocellular carcinoma growth and tumor angiogenesis in vivo. J Hepatol 2008; 49:977 - 984
- Whitehurst B, Flister MJ, Bagaitkar J, Volk L, Bivens CM, Pickett B, et al. Anti-VEGF-A therapy reduces lymphatic vessel density and expression of VEGFR-3 in an orthotopic breast tumor model. Int J Cancer 2007; 121:2181 - 2191
- Yu Y, Lee P, Ke Y, Zhang Y, Yu Q, Lee J, et al. A humanized anti-VEGF rabbit monoclonal antibody inhibits angiogenesis and blocks tumor growth in xenograft models. PLoS One 2010; 5:9072
- Zhang J, Lu A, Beech D, Jiang B, Lu Y. Suppression of breast cancer metastasis through the inhibition of VEGF-mediated tumor angiogenesis. Cancer Ther 2007; 5:273 - 286
- Ren B, Yee KO, Lawler J, Khosravi-Far R. Regulation of tumor angiogenesis by thrombospondin-1. Biochim Biophys Acta 2006; 1765:178 - 188
- Streit M, Velasco P, Brown LF, Skobe M, Richard L, Riccardi L, et al. Overexpression of thrombospondin-1 decreases angiogenesis and inhibits the growth of human cutaneous squamous cell carcinomas. Am J Pathol 1999; 155:441 - 452
- Kim HH, Kuwano Y, Srikantan S, Lee EK, Martindale JL, Gorospe M. HuR recruits let-7/RISC to repress c-Myc expression. Genes Dev 2009; 23:1743 - 1748
- Katsanou V, Papadaki O, Milatos S, Blackshear PJ, Anderson P, Kollias G, et al. HuR as a negative posttranscriptional modulator in inflammation. Mol Cell 2005; 19:777 - 789
- Khabar KS. The AU-rich transcriptome: more than interferons and cytokines and its role in disease. J Interferon Cytokine Res 2005; 25:1 - 10
- Khabar KS, Bakheet T, Williams BR. AU-rich transient response transcripts in the human genome: expressed sequence tag clustering and gene discovery approach. Genomics 2005; 85:165 - 175
- Meisner NC, Hackermuller J, Uhl V, Aszodi A, Jaritz M, Auer M. mRNA openers and closers: modulating AU-rich element-controlled mRNA stability by a molecular switch in mRNA secondary structure. Chembiochem 2004; 5:1432 - 1447
- Figueroa A, Cuadrado A, Fan J, Atasoy U, Muscat GE, Munoz-Canoves P, et al. Role of HuR in skeletal myogenesis through coordinate regulation of muscle differentiation genes. Mol Cell Biol 2003; 23:4991 - 5004
- van der Giessen K, Di-Marco S, Clair E, Gallouzi IE. RNAi-mediated HuR depletion leads to the inhibition of muscle cell differentiation. J Biol Chem 2003; 278:47119 - 47128
- Calaluce R, Gubin MM, Davis JW, Magee JD, Chen J, Kuwano Y, et al. The RNA binding protein HuR differentially regulates unique subsets of mRNAs in estrogen receptor negative and estrogen receptor positive breast cancer. BMC Cancer 2010; 10:126
- Ortega AD, Sala S, Espinosa E, Gonzalez-Baron M, Cuezva JM. HuR and the bioenergetic signature of breast cancer: a low tumor expression of the RNA-binding protein predicts a higher risk of disease recurrence. Carcinogenesis 2008; 29:2053 - 2061
- Ruhlen RL, Willbrand DM, Besch-Williford CL, Ma L, Shull JD, Sauter ER. Tamoxifen induces regression of estradiol-induced mammary cancer in the ACI. COP-Ept2 rat model. Breast Cancer Res Treat 2009; 117:517 - 524
- Smyth G. Gentleman RCV, Dudoit S, Irizarry R, Huber W. Limma: linear models for microarray data. Bioinformatics and computational Biology Solutions 2005; New York Springer
- Du P, Kibbe WA, Lin SM. lumi: a pipeline for processing Illumina microarray. Bioinformatics 2008; 24:1547 - 1548
- Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol 2004; 5:80
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society 1995; 57:289 - 300
- Maglott D, Ostell J, Pruitt KD, Tatusova T. Entrez Gene: gene-centered information at NCBI. Nucleic Acids Res 2005; 33:54 - 58
- Du P. lumiHumanAll.db: Illumina Human Expression BeadChips (include al versions: from version 1 to 3) annotation data 1.2.0 Rpv, Ed.
- Carlson M, Falcon S, Pages H, Li N. GO.db: A set of annotation maps describing the entire Gene Ontology 2.2.0 Rpv, Ed.
- Falcon S, Gentleman R. Using GOstats to test gene lists for GO term association. Bioinformatics 2007; 23:257 - 258
- Alexa A, Rahnenfuhrer J, Lengauer T. Improved scoring of functional groups from gene expression data by decorrelationg GO graph structure. Bioinformatics 2006; 22:1600 - 1607