Abstract
Chromosomal DNA must be precisely replicated in each cell cycle in order to ensure maintenance of genome stability. Most of the factors controlling this process have been identified in lower eukaryotes. Several factors involved in DNA replication are also important for the cell response to stress conditions. However, the regulation of DNA replication in multi-cellular organisms is still poorly understood. Using the Xenopus laevis egg cell-free system, we have recently identified a novel vertebrate protein named GEMC1 required for DNA replication. xGEMC1 is a Cyclin dependent kinase (CDK) target required the Cdc45 loading onto chromatin and it interacts with the checkpoint and replication factor TopBP1, which promotes its binding to chromatin during pre-replication complex formation. Here we discuss our recent findings and we propose possible roles for GEMC1. Interesting, recent studies have identified other proteins with analogous functions, showing a higher level of complexity in metazoan replication control compared to lower eukaryotes.
Introduction
In order for the genome to be faithfully maintained, chromosomal DNA must be precisely replicated and segregated in each cell cycle. Genome stability requires the coordination of DNA replication with cell cycle progression and DNA damage sensing and repair through mechanisms called checkpoints. Understanding the molecular details of these processes is extremely important since genomic instability, which arises from defects in their regulation, underpins aging, several human diseases and cancer predisposition.Citation1,Citation2
Eukaryotic DNA replication is a conserved process that starts with the assembly of a pre-replicative complex (pre-RC) at DNA replication origins in G1 phase. This event requires the sequential loading of the ORC1–6 complex together with Cdc6 and Cdt1 proteins onto chromatin, which lead to recruitment of the MCM2–7 helicase complex.Citation3 In S-phase, pre-RCs are then converted into bi-directional replication forks thanks to the activation of the helicase complexes. This step is mediated by two kinds of S-phase promoting kinases: Cdk2-CyclinE (S-phase CDK) and the Cdc7-Dbf4 kinase complex (DDK). In particular, phosphorylation events by CDK and DDK kinases promote the loading of the initiator factor Cdc45, in the presence of additional proteins including Mcm10, TopBP1 and the GINS complex. MCM2–7 complexes are then activated, leading to DNA unwinding and polymerase loading.Citation4
DNA replication has been studied in different model systems. Although the basic steps are conserved, many molecular details still need to be fully understood, especially in metazoans.
GEMC1: A Novel Vertebrate Protein Required for DNA Replication
In order to better understand the mechanisms of DNA replication in metazoans, we have recently looked for novel proteins containing known replicative motifs. We have identified a protein containing a coiled coil domain similar to Geminin, a well known factor that prevents MCM2–7 reloading onto fired origins, inhibiting Cdt1 activity.Citation5 Due to its structure, we called this protein GEMC1 (geminin coiled-coil containing protein 1). Our work showed that this protein is a novel factor required for chromosomal DNA replication in vertebrates.Citation6
We studied the function of GEMC1 in DNA replication mainly using the Xenopus egg extract system, which is able to recapitulate cell cycle progression in vitro.Citation7 We found that Xenopus GEMC1 (xGEMC1) binds chromatin in an ORC1–6 dependent manner at an early stage in DNA replication. To understand the role of xGEMC1, we have depleted xGEMC1 from egg extracts then monitored DNA replication of sperm nuclei. We found that, differently from Geminin, depletion of xGEMC1 inhibits DNA replication preventing Cdc45 from binding to chromatin without affecting binding of other factors like MCM2–7.Citation6 To better understand the role of xGEMC1 in this process we identified its binding partners. We found that xGEMC1 is able to interact directly with essential replication factors such as Cdc45 and the BRCT containing protein TopBP1, which is also required for Cdc45 chromatin loading.Citation8 Interestingly, the interaction with TopBP1 was found to be important for xGEMC1 recruitment onto chromatin during early stages of pre-RC formation. Taken together, these data suggest that a TopBP1-dependent recruitment of xGEMC1 is required for Cdc45 chromatin loading at the beginning of replication. Moreover, we found that xGEMC1 is able to interact with the kinase Cdk2-CyclinE both in vitro and in egg extracts. This interaction led us to investigate whether GEMC1, like many other replication factors, was a CDK substrate.Citation3 35S-labeled xGEMC1 incubated in egg extracts showed multiple phosphorylated forms roscovitine-sensitive. Although we have identified xGEMC1 Thr 153 as major Cdk2 phosphorylation site, we were able to suppress xGEMC1 phosphorylation only combining mutations of all eight CDK consensus sites. xGEMC1 phosphorylation is also inhibited by mutations in xGEMC1 R198NL cyclin-binding domain, which is required for the with Cdk2-CyclinE complex.Citation6 These results revealed that xGEMC1 is a strong CDK substrate. We finally investigated the functional meaning of this phosphorylation in Xenopus DNA replication. We discovered that replacement of endogenous xGEMC1 with recombinant xGEMC1 that could not be phosphorylated by Cdk2 (xGEMC1-8ST-A) was not able to restore DNA replication. Moreover, constitutively phosphorylated recombinant xGEMC1 (xGEMC1-8ST-D) was able to stimulate DNA replication, enhancing loading of Cdc45. Interestingly, xGEMC1 Cdk2-dependent phosphorylation is important but not essential for the interaction with TopBP1 as residual binding to TopBP1 in the absence of CDK activity was observed.Citation6 Overall, these findings suggest that xGEMC1 is a factor involved in the first steps of DNA replication, where it mediates the Cdk2- and TopBP1-dependent loading of Cdc45 onto chromatin, which is required for origin firing. As expected for an important factor involved in DNA replication, GEMC1 is highly conserved in vertebrates and close homologs can be found in different organisms including human. Preliminary data show that depletion of the GEMC1 mouse ortholog severely inhibits cell proliferation similar to Cdc45 depletion, suggesting a role for this factor also in mammalian DNA replication.Citation6 The challenge for the future is to understand how GEMC1 regulates DNA replication of mammalian cells.
CDK-Dependent Control of GEMC1 and its Role in DNA Replication
One of the most interesting finding is that xGEMC1's ability to stimulate DNA replication is dependent on its phosphorylation by Cdk2. The importance of CDK activity in the beginning of replication has been known for long time.Citation9 Together with Cdc7-Dbf4 (DDK), cyclin-dependent kinase is required for the activation of MCM2–7 helicase complexes in S phase. It is generally accepted that Cdc7-Dbf4 kinase phophorylates N-terminal tails of MCM2–7 in all eukaryotes, probably generating a conformational change that allows the interaction with Cdc45.Citation4 However, the main targets of CDK and their functions have been clearly identified only in yeast.
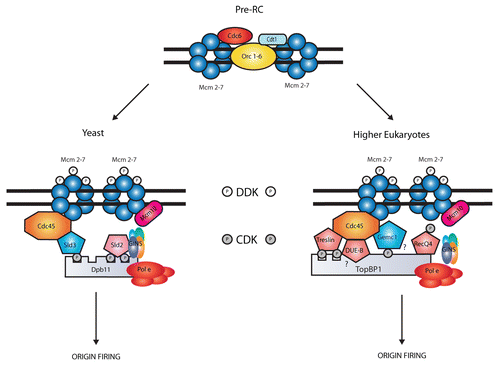
In budding yeast, two essential factors named Sld2 and Sld3 represent the minimal set of S-phase CDK substrates required for the initiation of DNA replication. Citation10,Citation11 Both factors were discovered as interactors of the four BRCT-containing protein Dpb11 (TopBP1/Mus101/Cut5 in metazoans), which is required for the recruitment of Cdc45 and GINS to the MCM2–7 complex at origins.Citation12,Citation13 The use of bypass mutants showed that Sld2 and Sld3 CDK-dependent phosphorylation is required for origin firing, stimulating the formation of complexes with the Dpb11 BRCTs (phosphopeptides binding domain).Citation10,Citation11,Citation14 The present model suggests that Sld3 interacts with Cdc45 and associates with origins in G1. After phosphorylation by CDK, Sld3 binds the Dpb11 N-terminal pair of BRCT repeats. At the same time, phosphorylated Sld2 interacts with the Dpb11 C-terminal pair of BRCT repeats leading to recruitment of GINS and DNA polymerase ε (, left).Citation15 While Dpb11 has well-established orthologs in metazoans, the presence of homologs Sld2 or Sld3 has been uncertain for many years. However, the CDK-dependent regulation of DNA replication is very conserved and functional Sld2/Sld3 homologs are expected to exist. Accordingly, CDK immunodepletion in Xenopus egg extracts prevents the initiation of DNA replication.Citation16
We showed that xGEMC1 represents an important CDK target involved in origin firing. Intriguingly xGEMC1 resembles Sld3 in three main features. First, xGEMC1 is able to interact directly with Cdc45 and TopBP1. Second, like Sld3, it is involved in Cdc45 recruitment onto replication origins. Third, CDK-dependent phosphorylation increases affinity for TopBP1 and stimulates both Cdc45 recruitment and origin firings.Citation6 Although no sequence similarity can be detected between Sld3 and xGEMC1, GEMC1 could represent a vertebrate Sld3 homolog (, right). However, it is also evident that the two factors differ in some aspects. For example, xGEMC1 CDK-dependent phosphorylation is not essential for the binding to TopBP1, although it enhances this interaction. Moreover, while Sld3 is required only for the beginning of replication, we detected xGEMC1 TopBP1-independent recruitment to chromatin after initiation of DNA replication, suggesting the possibility of additional binding partners of GEMC1 on DNA, not reported for Sld3. Finally, xGEMC1 phosphomimetic mutant (xGEMC1-8ST-D) is not able to fully restore DNA replication when CDK activity is inhibited by roscovitine in egg extract.Citation6 This last observation suggests that other CDK targets are required for replication initiation in Xenopus.
It is reasonable to think that another important CDK target could be an Sld2 functional homolog. Up to now, the unique protein proposed to have this kind of function is the RECQ helicase family member RecQ4.Citation17,Citation18 RecQ4 maintains the interaction with TopBP1 and is essential for DNA replication initiation in Xenopus egg extracts, promoting Polα loading. However in contrast with Sld2, this interaction and RecQ4 chromatin recruitment are not CDK-mediated in Xenopus, while in human CDK phosphorylation likely stimulates RecQ4 helicase activity, which would help the MCM2–7 complex in the DNA unwinding process.Citation19 These observations, together with the lack of sequence conservation suggest that particular replication factors such as Sld2 and Sld3 may have been subject to a rapid divergent evolution resulting in different origin firing controls in each species. This could be explained by the fact that these factors are not part of the replication machinery, but they simply mediate protein interactions for the beginning of replication and for this reason they could evolve more rapidly.Citation4 In addition, multicellular organisms might have developed a more complex regulation that requires more then two CDK substrates.Citation20 According to this hypothesis, it is interesting to notice how Dpb11 orthologs have acquired additional BRCT domains during evolution that could allow more interactions with phosphorylated proteins.Citation9 Overall, these observations agree with a possible scenario where multiple Sld2 or Sld3 homologs exist in metazoans, which suggests the existence of an Sld2/3-like protein family. Members of this family could have some sequence similarity with Sld2 and Sld3 but more important features should be: the ability to associate with TopBP1, a functional CDK-dependent phosphorylation and an important role in the initiation of DNA replication, promoting the recruitment of initiation factors like Cdc45 or GINS and the resulting DNA unwinding.
GEMC1 is a novel replication factor that fits perfectly in this category. Indeed, its ability to induce origin firing relies on its ability to induce Cdc45 recruitment onto chromatin. Moreover, it accomplishes this function in a CDK-dependent manner, working together with TopBP1.Citation6 Surprisingly, another two novel proteins have been recently identified and proposed as important CDK targets for the beginning of replication. Interestingly, both factors are able to interact with TopBP1.
The first was identified in Xenopus egg extracts as a TopBP1-binding partner and for this reason it was named Treslin (TopBP1-interacting, replication-stimulating protein).Citation21 Like yeast Sld3, it interacts with the first two N-terminal BRCT domains of TopBP1 and its depletion in egg extracts and human cells compromises DNA replication, abolishing Cdc45 chromatin recruitment. Even if evidence of direct phosphorylation is missing, Treslin appears to be a CDK target. However, Treslin and TopBP1 associate with chromatin independently. It has been proposed that Treslin phosphorylation by CDKs could only in a moment allow the interaction with TopBP1, which would promote the subsequent loading of Cdc45.Citation21 Moreover the Zebrafish analog of Treslin, called Ticrr (TopBP1-interacting checkpoint and replication regulator), has also been identified.Citation22 Taken together, these findings indicate that Treslin/Ticrr is another conserved essential CDK target in Metazoans.
The second protein that has been recently proposed as an important factor in beginning of DNA replication is DUE-B. First identified for its ability to bind DNA regions of predicted helical instability (DNA Unwinding Elements), the DUE-B protein has recently been shown to be necessary for replication initiation in Xenopus and mammalian systems.Citation23 Sequential immunoprecipitation showed that, like Treslin and GEMC1, DUE-B is able to interact with both Cdc45 and TopBP1, forming with them a ternary complex in vivo. Moreover, DUE-B binds replication origins in a pre-RC dependent way and in parallel with Cdc45. Immunodepletion of DUE-B in Xenopus extracts inhibits Cdc45 loading, strongly suggesting that DUE-B may function like Sld3. Finally, mass spectrometry analysis revealed that DUE-B is also phosphorylated. However, in contrast with Sld3, DUE-B phosphorylation is not essential for the interaction with TopBP1 but could act after the assembly of the TopBP1-DUE-B-Cdc45 complex with the pre-RC in order to strengthen the binding to Cdc45.Citation23
Overall, these observations suggest that in vertebrates the mechanism controlling initiation of chromosomal DNA replication is more complex compared to lower eukaryotes. While in budding yeast Sld2 and Sld3 are the unique CDK substrates required for origin firing, in metazoans four different factors (GEMC1, DUE-B, Treslin/Ticrr and RecQ4) have been proposed to act in an Sld2/3-like manner so far. Further studies are required to understand better the role of these new factors in DNA replication initiation and how they can work together. First of all, it is important to establish how every single factor interacts with the BRCT domains of TopBP1. It is known that among the 8 BRCTs present in the vertebrate homolog, the first three N-terminal repeats are essential for DNA replication.Citation21 It is reasonable to expect that all Sld2/3-like factors could interact with this region. In agreement with this, Treslin is able to interact with the first two N-terminal BRCTs of Xenopus TopBP1. How the other factors interact is still unknown. TopBP1's additional BRCT domains might mediate the interaction with GEMC1 and DUE-B, raising the possibility that these factors might function in complex with Treslin. Therefore a crucial aspect to investigate is the order in which these proteins are recruited to chromatin before fork establishment and verify the possibility that they can bind together to form functional complexes required for replication. Finally it is possible that additional CDK substrates with a role in DNA replication initiation would be identified in the future.
Possible Roles of GEMC1 in the Maintenance of Genome Stability
In order to maintain genome stability, all organisms must respond properly to endogenous/exogenous stress that tends to stall replication forks. When replication forks are stalled, eukaryotic cells activate a DNA damage checkpoint.Citation24 In S-phase, checkpoint mechanisms delay replication and prevent replication forks collapse to guarantee DNA synthesis restart.Citation1 For this reason DNA replication has to be highly coordinated with the DNA damage response. Indeed, replication structures are essential for the checkpoint activation, since they generate the primed single-stranded DNA (ssDNA), which represents the signal for ATR apical kinase activation. For this reason, several factors of the replication machinery are also involved in S-phase checkpoint activation. Some of them, such as MCM2–7 or Cdc45, are involved in the formation of those structures that signal during ongoing S-phase. Some others have additional checkpoint functions. One good example is TopBP1, which works not only in DNA replication initiation as previously described, but also as a direct activator of ATR following recruitment to the sites of replication stress.Citation22,Citation25 One of the main outputs of S-phase checkpoint is the inhibition of late replication origins firing in the presence of DNA damage. This could be achieved by preventing Cdc45 loading onto chromatin, for example inhibiting S-phase-promoting kinases.Citation24
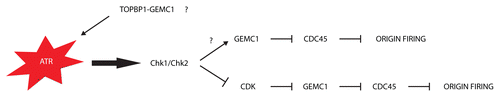
Our findings revealed GEMC1 as a strong CDK target and a TopBP1-interacting protein required for Cdc45 loading in vertebrates. Due to these features it is tempting to speculate that GEMC1 has a role also in response to replication stress. One possibility is that GEMC1 could be an S-phase checkpoint target controlling Cdc45 recruitment. Interestingly we have noticed that GEMC1 remains and accumulates on chromatin even after Cdc45 recruitment, suggesting the possibility of additional functions such as monitoring replication progression.Citation6 Another possibility is that GEMC1 could collaborate somehow with TopBP1 for the activation of checkpoint kinases (). Moreover, it is known that an ATM- and ATR-dependent checkpoint is involved in the regulation of origins firing even during an unperturbed cell cycle. In particular, replication intermediates and ssDNA generated from stochastically fired origins are able to trigger a DNA damage response that downregulates Cdk2 and Cdc7, preventing neighboring origins from firing.Citation26 This could lead to a decreased phosphorylation level of GEMC1, which would inhibit Cdc45 recruitment (). Future studies can be directed to define the role of GEMC1, as well as the role of other Sld2/3-like proteins, in the presence of replication stress.
GEMC1 represents an exciting novel factor to investigate in the future also for the development of anticancer therapies. Usually, chemotherapeutic drugs affect cancer cells replication inducing DNA damage and fork stalling. Another approach that could be adopted is to prevent replication restart of cancer cells, blocking the origin firing mechanism. This could be done by inhibiting S-phase-promoting kinases DDK or CDK or by inhibiting their targets such as GEMC1 or other Sld2/3-like proteins.Citation27
Conclusions
During evolution, eukaryotic cells have developed complex systems that allow replication of their large genome from multiple replication origins. Although the molecular mechanisms governing DNA replication initiation are generally conserved, differences between higher and lower eukaryotes exist. In particular, the requirement in vertebrates of additional factors controlled by phosphorylation events mediated by CDK is emerging. We have recently identified in GEMC1 a novel CDK target required for replication in metazoans. Further studies are required to better understand its regulation with other replication factors under normal and stressful conditions. These studies will clarify the role of these factors in promoting genome duplication and stability.
Figures and Tables
Figure 1 Replication origins are licensed after the loading of MCM2–7 helicase complexes. This process is conserved in all eukaryotes and requires the action of Cdt1 and Cdc6, which are recruited in G1-phase together with Orc1–6 (pre-RC). In S-phase, active CDKs and DDKs allow MCMs activation thanks to the recruitment of Cdc45 and other additional factors to the pre-RC. In yeast, Sld2 and Sld3 are the main CDK targets required for this step; they both interact with Dpb11 leading to Cdc45 recruitment and origin firing. In higher eukaryotes different Sld2/3 like-proteins (pentagons) are involved in this step. In particular, Gemc1 is phosphorylated by CDK and interacts with TopBP1 for the recruitment of Cdc45.
Acknowledgements
We thank members of the genome stability lab for their insightful comments. This work was funded by Cancer Research UK. V.C. is also supported by the European Research Council (ERC) start up grant (206281), the Lister Institute of Preventive Medicine and the EMBO Young Investigator Program (YIP).
References
- Aguilera A, Gómez-González B. Genome instability: a mechanistic view of its causes and consequences. Nat Rev Genet 2008; 9:204 - 217
- Negrini S, Gorgoulis VG, Halazonetis TD. Genomic instability-an evolving hallmark of cancer. Nat Rev Mol Cell Biol 2010; 11:220 - 228
- Diffley JFX. Regulation of early events in chromosome replication. Curr Biol 2004; 14:778 - 786
- Labib K. How do Cdc7 and cyclin-dependent kinases trigger the initiation of chromosome replication in eukaryotic cells?. Genes Dev 2010; 24:1208 - 1219
- Saxena S, Yuan P, Dhar SK, Senga T, Takeda D, Robinson H, et al. A dimerized coiled-coil domain and an adjoining part of geminin interact with two sites on Cdt1 for replication inhibition. Mol Cell 2004; 15:245 - 258
- Balestrini A, Cosentino C, Errico A, Garner E, Costanzo V. GEMC1 is a TopBP1-interacting protein required for chromosomal DNA replication. Nat Cell Biol 2010; 12:484 - 491
- Garner E, Costanzo V. Studying the DNA damage response using in vitro model systems. DNA Repair (Amst) 2009; 8:1025 - 1037
- Garcia V, Furuya K, Carr AM. Identification and functional analysis of TopBP1 and its homologs. DNA Repair 2005; 4:1227 - 1239
- Tanaka S, Tak YS, Araki H. The role of CDK in the initiation step of DNA replication in eukaryotes. Cell Division 2007; 2:16
- Zegerman P, Diffley JFX. Phosphorylation of Sld2 and Sld3 by cyclin-dependent kinases promotes DNA replication in budding yeast. Nature 2007; 445:281 - 285
- Tanaka S, Umemori T, Hirai K, Muramatsu S, Kamimura Y, Araki H. CDK-dependent phosphorylation of Sld2 and Sld3 initiates DNA replication in budding yeast. Nature 2007; 445:328 - 332
- Kamimura Y, Masumoto H, Sugino A, Araki H. Sld2, which interacts with Dpb11 in Saccharomyces cerevisiae, is required for chromosomal DNA replication. Mol Cell Biol 1998; 18:6102 - 6109
- Kamimura Y, Tak YS, Sugino A, Araki H. Sld3, which interacts with Cdc45 (Sld4), functions for chromosomal DNA replication in Saccharomyces cerevisiae. EMBO J 2001; 20:2097 - 2107
- Manke IA, Lowery DM, Nguyen A, Yaffe MB. BRCT repeats as phosphopeptide-binding modules involved in protein targeting. Science 2003; 302:636 - 639
- Muramatsu S, Hirai K, Tak YS, Kamimura Y, Araki H. CDK-dependent complex formation between replication proteins Dpb11, Sld2, Pol epsilon and GINS in budding yeast. Genes Dev 2010; 24:602 - 612
- Blow JJ, Nurse P. A cdc2-like protein is involved in the initiation of DNA replication in Xenopus egg extracts. Cell 1990; 62:855 - 862
- Matsuno K, Kumano M, Kubota Y, Hashimoto Y, Takisawa H. The N-terminal noncatalytic region of Xenopus RecQ4 is required for chromatin binding of DNA polymerase alpha in the initiation of DNA replication. Mol Cell Biol 2006; 26:4843 - 4852
- Sangrithi MN, Bernal JA, Madine M, Philpott A, Lee J, Dunphy WG, et al. Initiation of DNA replication requires the RECQL4 protein mutated in Rothmund-Thomson syndrome. Cell 2005; 121:887 - 898
- Xu X, Rochette PJ, Feyissa EA, Su TV, Liu Y. MCM10 mediates RECQ4 association with MCM2–7 helicase complex during DNA replication. EMBO J 2009; 28:3005 - 3014
- Errico A, Costanzo V. Differences in the DNA replication of unicellular eukaryotes and metazoans: known unknowns. EMBO Rep 2010; 11:270 - 278
- Kumagai A, Shevchenko A, Shevchenko A, Dunphy WG. Treslin collaborates with TopBP1 in triggering the initiation of DNA replication. Cell 2010; 140:349 - 359
- Sansam CL, Cruz NM, Danielian PS, Amsterdam A, Lau ML, Hopkins N, et al. A vertebrate gene, ticrr, is an essential checkpoint and replication regulator. Genes Dev 2010; 24:183 - 194
- Chowdhury A, Liu G, Kemp M, Chen X, Katrangi N, Myers S, et al. The DNA unwinding element binding protein DUE-B interacts with Cdc45 in preinitiation complex formation. Mol Cell Biol 2010; 30:1495 - 1507
- Zegerman P, Diffley JFX. DNA replication as a target of the DNA damage checkpoint. DNA Repair (Amst) 2009; 8:1077 - 1088
- Kumagai A, Lee J, Yoo HY, Dunphy WG. TopBP1 activates the ATR-ATRIP complex. Cell 2006; 124:943 - 955
- Shechter D, Costanzo V, Gautier J. ATR and ATM regulate the timing of DNA replication origin firing. Nat Cell Biol 2004; 6:648 - 655
- Swords R, Mahalingam D, O'Dwyer M, Santocanale C, Kelly K, Carew J, et al. Cdc7 kinase—a new target for drug development. Eur J Cancer 2010; 46:33 - 40