Abstract
S phase is characterized by the replication of DNA and assembly of chromatin. This requires the synthesis of large amounts of histone proteins to package the newly replicated DNA. Histone mRNAs are the only mRNAs that do not have polyA tails, ending instead in a conserved stemloop sequence. The stemloop binding protein (SLBP) that binds the 3' end of histone mRNA is cell cycle regulated and SLBP is required in all steps of histone mRNA metabolism. Activation of cyclin E/cdk2 prior to entry into S phase is critical for initiation of DNA replication and histone mRNA accumulation. At the end of S phase SLBP is rapidly degraded as a result of phosphorylation of SLBP by cyclin A/cdk1 and CK2 effectively shutting off histone mRNA biosynthesis. E2F1, which is required for expression of many S-phase genes, is regulated in parallel with SLBP and its degradation also requires a cyclin binding site, suggesting that it may also be regulated by the same pathway. It is likely that activation of cyclin A/cdk1 so helps inhibit both DNA replication and histone mRNA accumulation, marking the end of S phase and entry into G2 phase.
Progression through the cell cycle is driven by sequential activation of a series of protein kinases, the cyclin/cdks.Citation1 Activation of the G1 cyclins, cyclin D/cdk4/6 and then cyclin E/cdk2 result in the activation of cell growth pathways and then initiation of DNA replication. During S phase, cyclin A/cdk2 is essential for ongoing DNA replication. Prior to entry into mitosis, activation of cyclin B/cdk1 results in nuclear envelope breakdown. Completion of mitosis requires the inactivation of both mitotic cyclins, cyclin B/cdk1 and cyclin A/cdk1 by destruction of the cyclin subunits by the anaphase-promoting complex (APC). It is less clear whether there is a specific transition that occurs at the end of S phase and entry into G2-phase.
Histone protein synthesis is restricted to S phase and regulation of histone protein synthesis is accomplished by regulation of histone mRNA levels. Replication histone mRNAs have a unique structure since they are the only eukaryotic mRNAs that are not polyadenylated, ending instead in a conserved stemloop. A novel RNA-binding protein, stemloop binding protein (SLBP), binds the 3′ end of histone mRNA and participates in many aspects of histone mRNA metabolism.Citation2 SLBP is cell cycle-regulated and the protein is rapidly degraded at the end of S phase.Citation3,Citation4 In a recent study, we demonstrated that the degradation of SLBP, a protein that is limiting for histone mRNA accumulation, requires phosphorylation by cyclin A/cdk1 which in turn primes the phosphorylation of an adjacent threonine by casein kinase 2 (CK2).Citation5 SLBP is subsequently degraded by a still-unknown ubiquitin ligase. The degradation of SLBP effectively stops the accumulation of histone mRNA until the next cell cycle.Citation3 Here we show that a major S-phase transcription factor, E2F1, is regulated in parallel with SLBP, raising the possibility that the activation of cyclin A/cdk1 at the end of S phase may help stop both histone synthesis and DNA replication, just as the activation of cyclin E/cdk2 in late G1 provides the signal for both DNA replication and accumulation of histone mRNA.
Regulation of DNA Replication
The initiation of DNA replication in vertebrates has been studied extensively and clearly depends on activation of cyclin E/cdk2, which phosphorylates a number of target proteins leading to initiation of DNA replication. As in many biochemical pathways, there are likely cascades of kinases which ultimately modify many target proteins required for DNA replication. E2F1 is a critical transcription factor that regulates transcription of many genes encoding proteins required for DNA replication, including enzymes of deoxynucleotide metabolism, members of the Mcm and Orc complexes and components of the replication apparatus.Citation6 One set of cyclin E/cdk2 target proteins are members of the pRb family (pRb, p130 and p107).Citation7 Phosphorylation of these proteins results in their release from the transcription factor E2F1 and upregulation of transcription of these genes.
The regulation of initiation DNA replication has been extensively reviewed.Citation8–Citation10 Activation of cyclin E/cdk2 is required to initiate DNA replication, and a critical target of cyclin E/cdk2 is Cdc6. Phosphorylation of Cdc6 protects it from degradation by the APC allowing licensing of origins in late G1.Citation11,Citation12 Other direct targets of cyclin E/cdk2 include p27, an inhibitor of entry into S phase,Citation13 which is degraded prior to S-phase entry and cyclin E itself,Citation14 each of which are part of an autoregulatory loop. During S phase cyclin A/cdk2 remains active and is essential for continued S-phase progression.Citation15 The completion of DNA replication occurs only when every region of DNA has been replicated once and only once. Continued activity of cyclin A/cdk2 and cyclin A/cdk1 prevents origin relicensing and rereplication.Citation16 In the next G1 phase, the cell must then license the origins so they can be fired again.Citation9
Cyclin/cdks and Histone Protein Synthesis
The replication of DNA is accompanied by the immediate assembly of the newly replicated DNA into chromatin. Thus, at the same time that DNA replication starts, there must be a rapid increase in the rate of histone protein synthesis. When DNA replication is complete, histone-protein accumulation must stop quickly. The regulation of histone protein synthesis is mediated by rapid changes in histone mRNA, as well as degradation of excess histone protein.Citation17,Citation18 Accumulation of histone mRNA is a result of an increase in the rate of histone gene transcription and the efficiency of histone pre-mRNA processing.Citation19 The molecular mechanisms that lead to activation of histone mRNA expression are not completely understood. However, the activity of cyclin E/cdk2 is required and an essential cyclin E substrate is the protein NPAT,Citation20–Citation22 which is found in the Histone Locus bodies. These contain factors required for histone mRNA biosynthesis and are adjacent to the histone genes.Citation23,Citation24 Phosphorylation of NPAT by cyclin E/cdk2 is necessary for accumulation of histone mRNA.Citation25 Thus, cyclin E/cdk2 is likely at the top of a cascade that results in both initiation of DNA replication as a result of phosphorylation of cdc6 and accumulation of histone mRNAs as a result of phosphorylation of NPAT.
At the end of S phase, histone mRNA is rapidly degraded and histone mRNA biosynthesis is inhibited. The degradation of histone mRNA is directly regulated by changes in the rate of DNA replication, in response to the demand for histone protein to package newly replicated DNA.Citation26,Citation27 A critical protein for histone mRNA metabolism is the SLBP, which binds to the 3′ end of histone mRNA and participates in histone pre-mRNA processing as well as histone mRNA degradation. SLBP is cell cycle regulated and the protein is present only in S-phase cells.Citation4 Since histone gene transcription continues outside of S phase at a reduced rate,Citation28 rapid degradation of SLBP at the end of S phase is necessary to shut off histone mRNA biosynthesis, by preventing histone pre-mRNA processing.Citation3 Nuclear extracts from G1- or G2-phase cells do not process histone mRNA, but the addition of recombinant SLBP restores processing.Citation3 Accumulation of SLBP just prior to entry into S phase is essential to allow the accumulation of histone mRNA necessary for histone protein synthesis. SLBP degradation is the result of phosphorylation of two threonine residues (T60 and 61) and also requires a putative cyclin binding site.Citation3 Thus regulation of SLBP levels is the critical component of postranscriptional regulation of histone mRNA levels ().
E2F1 is Regulated in Parallel with SLBP
E2F1 has been reported to be a cell cycle regulated protein,Citation29 although the elements in E2F1 responsible for cell cycle regulation have not been defined. A diagram of E2F1 with the domains that have been previously identified is shown in . These include a DNA-binding domain; a heterodimerization domain that interacts with DP, the E2F1 partner;Citation30 a domain that binds Rb and is required for stabilization of E2F1 when Rb is overexpressed;Citation31–Citation33 a domain that binds the F-box protein Skp2 that has been proposed to mediate E2f1 degradation as part of the cul1 E3 ligase;Citation29 an Arf binding site also involved in E2F degradationCitation34 and a domain that contains a cyclin A binding site.Citation35,Citation36
We confirmed that E2F1 is cell cycle regulated in HeLa cells and that the levels of E2F1 protein parallel the levels of SLBP, increasing as cells enter S phase and decreasing at the end of S phase, suggesting that E2F1, specifically, is degraded at the end of S phase (, lanes 5 and 6). E2F1 accumulates in parallel with SLBP as cells approach the next S phase. Similar results were obtained with E2F2 and E2F3 (Dong J and Marzluff WF, unpublished). Treatment of mid-S-phase cells with MG132 prevented the degradation of both SLBP and E2F1 (, lane 5). To determine which regions of E2F1 were required for degradation of E2F1 at the end of S phase we made a number of deletions in E2F1 and created cell lines expressing the mutant E2F1 containing an N-terminal HA tag. We deleted a variety of regions previously reported to be involved in degradation of E2F1 under a variety of conditions. These include the SKP2 binding site (amino acids 1–42),Citation29 the pRB binding site (amino acids 409–426)Citation37,Citation38 and the ARF binding site (amino acids 426–437).Citation34 The full-length HA-tagged protein was regulated appropriately (, top) and a mutant (aa 42–379) that removed the Skp2 binding site, the pRb binding site and the Arf binding site was also degraded at the same time as SLBP (, middle). This result indicated that the previously characterized regions that affected E2F1 stability in different conditions were not required for degradation of E2F1 at the end of S phase. Further deletion of the aminoterminus that removed amino acids 1–107 (108–437) and removed the cyclin A binding site resulted in stabilization of the E2F1 protein at the end of S phase (, bottom).
Additional deletions of either amino acids 109–190 or 191–299 from the center of the E2F1 protein had no effect on E2F1 degradation at the end of S phase (, top and middle). However, a mutant E2F1 with amino acids 300–379 deleted was not degraded at the end of S phase (, bottom), demonstrating that there were two regions of E2F1 required for degradation at the end of S phase; a sequence between amino acids 43 and 108 and the region between amino acids 300 and 379 ().
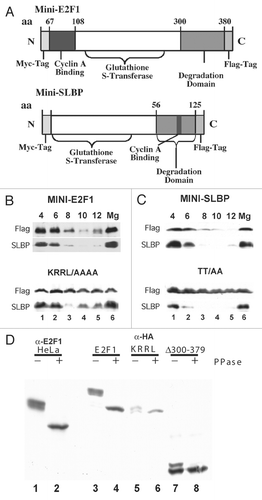
To demonstrate that these two regions were sufficient for degradation of E2F1 we created a miniE2F1 gene with GST substituted for the region between amino acids 49–107 and 300–380 (). The region from 67–108 contains the previously described cyclin binding site,Citation35 with the core of the cyclin binding siteCitation36,Citation39 at amino acids 89–92 of E2F1. We mutated the core region KRRL to AAAA in the E2F minigene. As a control we also used the SLBP mini gene,Citation5 which contained aa 56–125 fused to GST (). This region contains both the cyclin binding site and the SFTTP phosphorylation site. We mutated the two threonines in the phosphorylation site to alanines (SLBP-AA). Stable cell lines were created expressing each of these minigenes. Each of the minigenes contained a myc-tag at the N terminus and a FLAG-tag at the C terminus. The wild-type E2F minigene was degraded at the end of S phase (, top), while the mutant E2F minigene was stable throughout S phase and G2 phase (, bottom). Similar results were obtained for the SLBP minigenes () as previously reported.Citation5 Thus both SLBP and E2F1 contain a cyclin binding site that is required for their degradation at the end of S phase.
The region between aa 300 and 379 of E2F1 contains numerous SP and TP sites, suggesting that one or more of these sites is phosphorylated to trigger degradation of E2F1. To demonstrate the phosphorylation of E2F1, we treated lysat es with lambda protein phosphatase and resolved the lysates on a long polyacrylamide gel, using anti-E2F1 to detect the endogenous protein and anti-HA to detect the exogeous protein. Endogenous E2F1 was quantitatively phosphorylated in S-phase cells, and was detected as a broad band (, lane 1). Treatment with phosphatase converted E2F1 to single species (, lane 2). We analyzed HA-tagged full-length E2F1, the mutation of the core cyclin binding site and the deletion of amino acids 370–379. Similar results to the endogenous E2F1 were obtained with the HA-tagged full-length protein (, lanes 3 and 4). Most of the phosphorylation was dependent on the cyclin binding site (, lanes 5 and 6). Mutation of the cyclin binding site resulted in converting E2F1 to two components, one with identical migration to the phosphatase-treated protein and one migrating slightly slower, suggesting there is a single phosphorylation site that did not depend on the cyclin binding site. The deletion of aa 300–379 gave a similar result to mutating the cyclin binding site. E2F migrated as a doublet, the lower band of which migrated identically with phosphatase treated E2F1 (, lanes 7 and 8). We conclude that the cyclin binding site is required for bulk of the phosphorylation of E2F1, which is found on amino acids 300–379. The bulk phosphorylation pattern of E2F1 is similar in early and late S phase, suggesting that there are multiple cyclin-dependent phosphorylations on E2F1. Due to the multiple SP and TP sequences in this region, we have not been able to identify the specific phosphorylation sites on E2F1.
Cyclin A/cdk1 is Required for the Degradation of SLBP at the End of S phase
Our recent studyCitation5 demonstrates that the critical phosphorylation event for SLBP degradation is carried out by cyclin A/cdk1, a kinase which is activated at late S phase.Citation40 Recombinant cyclin A/cdk1 phosphorylates T61 of SLBP but recombinant cyclin A/cdk2 does not. Degradation of SLBP requires a KRKL sequence about 40 amino acids after the phosphorylation sites and phosphorylation of T61 of SLBP by cyclin A/cdk1 depends on the KRKL sequence. Given the requirement of the cyclin A binding site for E2F1 degradation () and the timing of the degradation, it is likely that cyclin A/cdk1 phosphorylates a critical site on E2F1 leading to its degradation.
The second kinase required for degradation of SLBP is casein kinase II (CK2). CK2 phosphorylates T60 of SLBP, but only after T61 has been phosphorylated by cyclin A/cdk1.Citation5 CK2 has also been shown to be important for proper cell cycle progression. CK2 activity is required for the G1/S and G2/M transition in yeastCitation41 and mammalian cells.Citation42,Citation43 The consensus CK2 target site requires nearby negative charges and often requires priming phosphorylations by other kinases. Another known example of cooperation of CK2 and cyclin/cdk1 is in G2, when phosphorylation of Wee1 by a cyclin/Cdk1 primes phosphorylation by CK2 resulting in production of a phosphodegron that is necessary for proper Wee1 degradation and proper G2/M transition.Citation43
Cyclin A/cdk1 and DNA Replication
A number of substrates for cyclin A/cdk1 have been identified recently and many of these share a property that phosphorylation contributes to the inactivation of proteins required for DNA replication. These include Orc1Citation44 and Fen1,Citation45 each of which is required for DNA replication, and the CDP/Cux transcription factor,Citation46,Citation47 which is necessary for transcription of the DNA polymerase á gene. These results are consistent with the possibility that activation of cyclin A/cdk1 at the end of S phase is an event that marks the end of S phase. Cyclin A/cdk1 may also help prevent premature activation of the APC during G2 by phosphorylating cdh1 on the APC.Citation40 The other known substrate of cyclin A/cdk1 is cdc25B,Citation48 which is degraded as a result of phosphorylation by cyclin A/cdk1, possibly as part of an autoregulatory loop. Thus, one role of cyclin A/cdk1 may be to irreversibly inactivate DNA replication to prevent re-replication of DNA, as well as inactivating histone mRNA biosynthesis at the end of S phase.
There are likely other examples of phosphorylations that might be carried out by cyclin A/cdk1 but have only been identified as cyclin A substrates. For example, a cyclin A/cdk is believed to have role in cyclin B/cdk1 activation,Citation49 another event that occurs at G2. It is possible that cyclin A/cdk1 activity may be needed for the initial, activating phosphorylation of Cdc25 and/or inhibitory phosphorylation of Wee1 to trigger the cyclin B/cdk1 auto-activation loop for robust cyclin B/cdk1 activation and M-phase entry.
The mechanism by which the cell modulates cyclin A/cdk1 and cyclin A/cdk2 activity remains elusive since all three polypeptides are present throughout S and G2-phase, until cyclin A is degraded at mitosis. Recent studies by Merrick and colleagues show that cyclin A binds cdk2 early in S phase and only binds cdk1 near the end of S phase. They also point out that the different activation pathways for cdk1 and cdk2 contribute to the initation of cyclin A/cdk1 activity at the end of S phase.Citation52,Citation53 In addition chk1 kinase acts to prevent premature activation of cyclin A/cdk1, by destabilizing Cdc25A.Citation52,Citation53 It is possible that cyclin A-bound cdk1 is either more readily dephosphorylated by Cdc25 and/or resistant to inhibitory phosphorylations by kinases such as Wee1. This may be the reason for detection of cyclin A/cdk1 activity starting from late S phase toward G2, where it peaks, while cyclin B/cdk1 activity stays low until G2/M when a rapid activity increase causes M phase entry.
Conclusions
The major cell cycle transitions are driven by changes in activities of specific cyclin/cdks. Our results together with those of others, suggest that the transition from S phase to G2 phase may be mediated by the increase in the activity of cyclin A/cdk1 at S/G2. This occurs prior to the robust activation of cyclin B/cdk1, which results in nuclear envelope breakdown and entry into mitosis. Activation of cyclin A/cdk1 results in inhibition of histone mRNA synthesis due to degradation of SLBP, as well as degradation of E2F1-3 and inhibition of several components involved in DNA replication. Thus activation of cyclin A/cdk1 helps shut down the chromosome replication machinery and plays a critical role as the cell transitions from S phase into G2 phase when it prepares to enter mitosis.
Figures and Tables
Figure 1 Regulation of histone mRNA during the mammalian cell cycle. The changes in levels of histone mRNA, SLBP and SLBP mRNA during the cell cycle are shown. Cyclin E/cdk2 activation leads to phosphorylation of NPAT and likely translation of SLBP mRNA resulting in activation of histone mRNA accumulation. At the end of S phase, cessation of DNA replication results in degradation of histone mRNA, while activation of cyclin A/cdk1 is required for SLBP degradation.
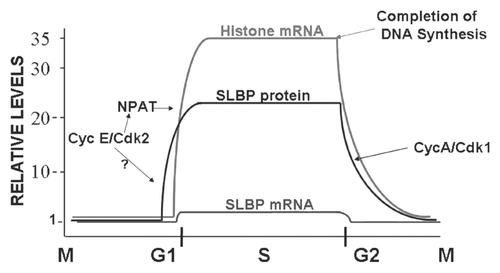
Figure 2 Definition of a region required for E2F1 degradation at the end of S phase. (A) A schematic of the domains of the E2F1 protein is shown. (B and C) HeLa cells were synchronized by double-thymidine block and released into S phase. At the indicated times, lysates were prepared and analyzed for SLBP and E2F1 levels by western blotting. In (C) MG132 was added 4 hrs after release into S phase and lysates prepared 6 hrs later (lane 5). (D and E) HeLa cells stably expressing the E2F1 constructs tagged with HA (shown in F) on the amino terminus were synchronized by double-thymidine block; lysates prepared and analyzed by western blotting for the HA-tagged protein,SLBP and where the tagged protein was resolved from the wild-type protein, for endogenous E2F1. In (D), the top part is full-length E2F1, the middle part deletion of amino acids 1–42 and the bottom part, deletion of amino acids 1–107. The bottom part was probed with anti-E2F1 and the endogenous and mutant protein were detected. In (E), the top part is deletion of amino acids 109–190, the middle part deletion of amino acids 191–299 and the bottom part deletion of amino acids 300–379. This blot was probed with both anti-E2F1 and anti-HA. (F) A summary of the effect of the deletions on degradation of E2F1 at the end of S phase is shown.
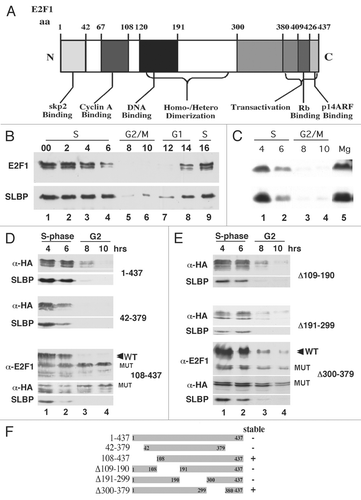
Figure 3 Phosphorylation of E2F1 requires a cyclin binding site for E2F1 degradation. (A) The indicated minigenes for SLBP5 and E2F1 are shown. Amino acids 56–125 of SLBP are sufficient to direct degradation of the protein. The E2F minigene contains the two fragments of E2F1 identified in , 67–108 and 300–380 separated by GST. Point mutants of the core of the cyclin binding site of E2F1 and in the TT which is the target of phosphorylation of SLBP were also constructed in the context of the minigene. (B and C) Cells stably expressing the E2F1 minigene and mutant minigene (B) or the SLBP minigene or mutant minigene (C) were synchronized by double-thymidine block, released into S phase. Parallel cultures expressing the SLBP and E2F minigenes were synchronized and lysates were prepared at the indicated times after release and analyzed by western blotting for the FLAG-tag on the minigene or SLBP antibody to detect endogenous SLBP. (D) Cells expressing HA-tagged wild-type E2F1, the cyclin binding site mutant in the wild-type protein, of the 300–379 deletion (lanes 3–8) were synchronized by double-thymidine block and lysate prepared 4 hrs after release into S phase. A portion of the lysate was treated with lambda protein phosphatase. Equal amounts of the treated and untreated lysates were analyzed by western blotting using the anti-HA antibody after resolution of long polyacrylamide gels for an extended amount of time to resolve phosphorylated forms. The endogenous E2F1 was analyzed in lanes 1 and 2 using the E2F1 antibody. Note the difference in mobility of the endogenous E2F1 and the tagged-E2F1 is due to the HA-tag.
Acknowledgements
This work was supported by NIH grant GM29832 to W.F.M. We thank Jean Cook for critical comments on the manuscript.
References
- Woo RA, Poon RY. Cyclin-dependent kinases and S-phase control in mammalian cells. Cell Cycle 2003; 2:316 - 324
- Marzluff WF, Wagner EJ, Duronio RJ. Metabolism and regulation of canonical histone mRNAs: life without a poly(A) tail. Nat Rev Genet 2008; 9:843 - 854
- Zheng LX, Dominski Z, Yang X, Elms P, Raska CS, Borchers CH, et al. Phosphorylation of SLBP on two threonines triggers degradation of SLBP, the sole cell cycle regulated factor required for regulation of histone mRNA processing, at the end of S-phase. Mol Cell Biol 2003; 23:1590 - 1601
- Whitfield ML, Zheng LX, Baldwin A, Ohta T, Hurt MM, Marzluff WF. Stem-loop binding protein, the protein that binds the 3′ end of histone mRNA, is cell cycle regulated by both translational and posttranslational mechanisms. Mol Cell Biol 2000; 20:4188 - 4198
- Koseoglu MM, Graves LM, Marzluff WF. Phosphorylation of threonine 61 by cyclin A/Cdk1 triggers degradation of stemloop binding protein at the end of S phase. Mol Cell Biol 2008; 28:4469 - 4479
- Bracken AP, Ciro M, Cocito A, Helin K. E2F target genes: Unraveling the biology. Trends Biochem Sci 2004; 29:409 - 417
- Harbour JW, Luo RX, Santi AD, Postigo AA, Dean DC. Cdk phosphorylation triggers sequential intramolecular interactions that progressively block Rb functions as cells move through G1. Cell 1999; 98:859 - 869
- Diffley JF. Regulation of early events in chromosome replication. Curr Biol 2004; 14:778 - 786
- Machida YJ, Hamlin JL, Dutta A. Right place, right time and only once: Replication initiation in metazoans. Cell 2005; 123:13 - 24
- Sclafani RA, Holzen TM. Cell cycle regulation of DNA replication. Annu Rev Genet 2007; 41:237 - 280
- Mailand N, Diffley JF. CDKs promote DNA replication origin licensing in human cells by protecting Cdc6 from APC/C-dependent proteolysis. Cell 2005; 122:915 - 926
- Duursma A, Agami R. p53-Dependent regulation of Cdc6 protein stability controls cellular proliferation. Mol Cell Biol 2005; 25:6937 - 6947
- Sheaff RJ, Groudine M, Gordon M, Roberts JM, Clurman BE. Cyclin E-CDK2 is a regulator of p27Kip1. Genes Dev 1997; 11:1464 - 1478
- Welcker M, Singer J, Loeb KR, Grim J, Bloecher A, Gurien-West M, et al. Multisite phosphorylation by Cdk2 and GSK3 controls cyclin E degradation. Mol Cell 2003; 12:381 - 392
- Pagano M, Pepperkok R, Verde F, Ansorge W, Draetta G. Cyclin A is required at two points in the human cell cycle. EMBO J 1992; 11:961 - 971
- Hochegger H, Dejsuphong D, Sonoda E, Saberi A, Rajendra E, Kirk J, et al. An essential role for Cdk1 in S-phase control is revealed via chemical genetics in vertebrate cells. J Cell Biol 2007; 178:257 - 268
- Gunjan A, Verreault A. A Rad53 kinase-dependent surveillance mechanism that regulates histone protein levels in S. cerevisiae. Cell 2003; 115:537 - 549
- Gunjan A, Paik J, Verreault A. The emergence of regulated histone proteolysis. Curr Opin Genet Dev 2006; 16:112 - 118
- Harris ME, Böhni R, Schneiderman MH, Ramamurthy L, Schümperli D, Marzluff WF. Regulation of histone mRNA in the unperturbed cell cycle: Evidence suggesting control at two posttranscriptional steps. Mol Cell Biol 1991; 11:2416 - 2424
- Zhao JY, Dynlacht B, Imai T, Hori T, Harlow E. Expression of NPAT, a novel substrate of cyclin E-CDK2, promotes S-phase entry. Genes Dev 1998; 12:456 - 461
- Ma TL, Van Tine BA, Wei Y, Garrett MD, Nelson D, Adams PD, et al. Cell cycle-regulated phosphorylation of p220NPAT by cyclin E/cdk2 in Cajal bodies promotes histone gene transcription. Genes Dev 2000; 14:2298 - 2313
- Zhao JY, Kennedy BK, Lawrence BD, Barbie DA, Matera AG, Fletcher JA, et al. NPAT links cyclin E-Cdk2 to the regulation of replication-dependent histone gene transcription. Genes Dev 2000; 14:2283 - 2297
- Ghule PN, Dominski Z, Yang XC, Marzluff WF, Becker KA, Harper JW, et al. Staged assembly of histone gene expression machinery at subnuclear foci in the abbreviated cell cycle of human embryonic stem cells. Proc Natl Acad Sci USA 2008; 44:16964 - 16969
- Liu JL, Buszczak M, Gall JG. Nuclear bodies in the Drosophila germinal vesicle. Chromosome Res 2006; 14:465 - 475
- Ye X, Wei Y, Nalepa G, Harper JW. The cyclin E/Cdk2 substrate p220(NPAT) is required for S-phase entry, histone gene expression and Cajal body maintenance in human somatic cells. Mol Cell Biol 2003; 23:8586 - 8600
- Sariban E, Wu RS, Erickson LC, Bonner WM. Interrelationships of protein and DNA syntheses during replication of mammalian cells. Mol Cell Biol 1985; 5:1279 - 1286
- Kaygun H, Marzluff WF. Regulated degradation of replication-dependent histone mRNAs requires both ATR and Upf1. Nat Struct Mol Biol 2005; 12:794 - 800
- DeLisle AJ, Graves RA, Marzluff WF, Johnson LF. Regulation of histone mRNA production and stability in serum stimulated mouse fibroblasts. Mol Cell Biol 1983; 3:1920 - 1929
- Marti A, Wirbelauer C, Scheffner M, Krek W. Interaction between ubiquitin-protein ligase SCFSKP2 and E2F-1 underlies the regulation of E2F-1 degradation. Nat Cell Biol 1999; 1:14 - 19
- Jordan KL, Haas AR, Logan TJ, Hall DJ. Detailed analysis of the basic domain of the E2F1 transcription factor indicates that it is unique among bHLH proteins. Oncogene 1994; 9:1177 - 1185
- Hofmann F, Martelli F, Livingston DM, Wang ZY. The retinoblastoma gene product protects E2F-1 from degradation by the ubiquitin-proteasome pathway. Genes Dev 1996; 10:2949 - 2959
- Hateboer G, Kerkhoven RM, Shvarts A, Bernards R, Beijersbergen RL. Degradation of E2F by the ubiquitin-proteasome pathway: Regulation by retinoblastoma family proteins and adenovirus transforming proteins. Genes Dev 1996; 10:2960 - 2970
- Campanero MR, Flemington EK. Regulation of E2F through ubiquitin-proteasome-dependent degradation: Stabilization by the pRB tumor suppressor protein. Proc Natl Acad Sci USA 1997; 94:2221 - 2226
- Martelli F, Hamilton T, Silver DP, Sharpless NE, Bardeesy N, Rokas M, et al. p19ARF targets certain E2F species for degradation. Proc Natl Acad Sci USA 2001; 98:4455 - 4460
- Xu M, Sheppard KA, Peng CY, Yee AS, Piwnica-Worms H. Cyclin A/CDK2 binds directly to E2F-1 and inhibits the DNA-binding activity of E2F-1/DP-1 by phosphorylation. Mol Cell Biol 1994; 14:8420 - 8431
- Adams PD, Sellers WR, Sharma SK, Wu AD, Nalin CM, Kaelin WG Jr. Identification of a cyclin-cdk2 recognition motif present in substrates and p21-like cyclin-dependent kinase inhibitors. Mol Cell Biol 1996; 16:6623 - 6633
- Helin K, Harlow E, Fattaey A. Inhibition of E2F-1 transactivation by direct binding of the retinoblastoma protein. Mol Cell Biol 1993; 13:6501 - 65018
- Flemington EK, Speck SH, Kaelin WG Jr. E2F-1-mediated transactivation is inhibited by complex formation with the retinoblastoma susceptibility gene product. Proc Natl Acad Sci USA 1993; 90:6914 - 6918
- Russo AA, Jeffrey PD, Patten AK, Massagué J, Pavletich NP. Crystal structure of the p27Kip1 cyclin-dependent-kinase inhibitor bound to the cyclin A-cdk2 complex. Nature 1996; 382:325 - 331
- Mitra J, Enders GH, Azizkhan-Clifford J, Lengel KL. Dual regulation of the anaphase promoting complex in human cells by cyclin A-Cdk2 and cyclin A-Cdk1 complexes. Cell Cycle 2006; 5:661 - 666
- Glover CV III. On the physiological role of casein kinase II in Saccharomyces cerevisiae. Prog Nucleic Acid Res Mol Biol 1998; 59:95 - 133
- Pepperkok R, Lorenz P, Ansorge W, Pyerin W. Casein kinase II is required for transition of G0/G1, early G1 and G1/S-phases of the cell cycle. J Biol Chem 1994; 269:6986 - 6991
- Watanabe N, Arai H, Iwasaki J, Shiina M, Ogata K, Hunter T, et al. Cyclin-dependent kinase (CDK) phosphorylation destabilizes somatic Wee1 via multiple pathways. Proc Natl Acad Sci USA 2005; 102:11663 - 11668
- Li CJ, Vassilev A, DePamphilis ML. Role for Cdk1 (Cdc2)/cyclin A in preventing the mammalian origin recognition complex's largest subunit (Orc1) from binding to chromatin during mitosis. Mol Cell Biol 2004; 24:5875 - 5886
- Henneke G, Koundrioukoff S, Hubscher U. Phosphorylation of human Fen1 by cyclin-dependent kinase modulates its role in replication fork regulation. Oncogene 2003; 22:4301 - 4313
- Santaguida M, Ding Q, Berube G, Truscott M, Whyte P, Nepveu A. Phosphorylation of the CCAAT displacement protein (CDP)/Cux transcription factor by cyclin A-Cdk1 modulates its DNA binding activity in G(2). J Biol Chem 2001; 276:45780 - 45790
- Santaguida M, Nepveu A. Differential regulation of CDP/Cux p110 by cyclin A/Cdk2 and cyclin A/Cdk1. J Biol Chem 2005; 280:32712 - 32721
- Baldin V, Cans C, Knibiehler M, Ducommun B. Phosphorylation of human CDC25B phosphatase by CDK1-cyclin A triggers its proteasome-dependent degradation. J Biol Chem 1997; 272:32731 - 32734
- Fung TK, Ma HT, Poon RY. Specialized roles of the two mitotic cyclins in somatic cells: cyclin A as an activator of M phase-promoting factor. Mol Biol Cell 2007; 18:1861 - 1873
- Merrick KA, Fisher RP. Putting one step before the other: distinct activation pathways for Cdk1 and Cdk2 bring order to the mammalian cell cycle. Cell Cycle 2010; 9:706 - 714
- Merrick KA, Larochelle S, Zhang C, Allen JJ, Shokat KM, Fisher RP. Distinct activation pathways confer cyclin-binding specificity on Cdk1 and Cdk2 in human cells. Mol Cell 2008; 32:662 - 672
- Nakanishi M, Katsuno Y, Niida H, Murakami H, Shimada M. Chk1-cyclin A/Cdk1 axis regulates origin firing programs in mammals. Chromosome Res 2010; 18:103 - 113
- Katsuno Y, Suzuki A, Sugimura K, Okumura K, Zineldeen DH, Shimada M, et al. Cyclin A-Cdk1 regulates the origin firing program in mammalian cells. Proc Natl Acad Sci USA 2009; 106:3184 - 3189