Abstract
In our recent paper, we identified a TIR encoding gene which is required for resistance against a broad range of necrotrophic fungi. Here we present this finding in a broader perspective and discuss the unique features of this gene which might explain its role as a general regulator of resistance responses against a class of pathogens that have previously not been associated to the classical resistance (R) gene type of defense.
Plant Defense to Necrotrophic Fungal Pathogens
An organism that kills host tissue, live and reproduce on energy derived from dead cells is termed a necrotroph. A life style that is opposite to a biotroph, which require living cells for successful colonization. Necrotrophic fungal pathogens are responsible for some of the most severe crop failures and have through epidemics caused the deaths of millions throughout history.Citation1,Citation2 With an increased world demand for food and other plant-derived agricultural products, which is currently illustrated by skyrocketing food prices,Citation3 the issue of plant disease is as pressing as ever. Despite this, comparatively little is known about resistance towards necrotrophic fungi as compared to the frequently used biotrophic oomycete model Hyaloperonospera parasitica and the hemibiotrophic bacterial model Pseudomonas syringae on Arabidopsis. The latter having a biotrophic lifestyle when it initially invades the plant, but later turn necrotrophicCitation4. Until recently, it was envisaged that only biotrophic pathogens were subject to gene-for-gene type of resistance interactions leading to a salicylic acid (SA) dependent response, whereas the resistance towards necrotrophs was mainly based on phytotoxin production and a set of defense responses requiring jasmonic acid (JA) and ethylene (ET) and mostly regulated by complex traits.Citation5–Citation7 However, Leptosphaeria maculans, a hemibiotrophic fungal pathogen causing devastating crop losses,Citation8 has a primarily necrotrophic nature but shows a clear gene-for-gene relationship with its Brassica hosts. This fungus is closely related to the necrotrophic Alternaria species. Interestingly, L. maculans resistance in Arabidopsis was recently found to require dominant resistance (R) genes of the same protein structure (TIR-NB-LRR) as those required for resistance against some biotrophs.Citation9 It is very likely that the gene-for-gene relationships between L. maculans and Brassica species depend on the same family of R genes,Citation10 but this has yet to be shown. On the other hand, Arabidopsis, in contrast to B. napus, shows a strong R gene independent pre-invasion response indicating that the Arabidopsis-L. maculans model borders that of a non-host interaction.Citation11 Further challenges towards the simple division between a SA-dependent response towards biotrophs and a JA/ET dependent response towards necrotrophs was also illustrated via L. maculans, where all three hormones play little or no significant role in resistance while a hormone primarily associated to abiotic stress, abscisic acid (ABA), was found to be required for the R gene dependent resistance against this pathogen.Citation12,Citation13 The unexpected finding that a short relative of the TIR-NB-LRR family of disease resistance genes is responsible for resistance against a wide range of necrotrophic fungi further complicates the picture, since no R gene pathway mutants have been clearly associated to any of the major fungal necrotroph models (A. brassicicola, Botrytis cinerea and Plectosphaerella cucumerina) on Arabidopsis.Citation14 Here we describe the recently identified gene resistance to Leptosphaeria maculans 3, RLM3, and the unique features of this gene, which may explain its broad-range effect against a truly devastating class of pathogens.
Identification and Cloning of RLM3
In a screening for L. maculans susceptible Arabidopsis accessions, only one out of 168 tested showed a clear susceptible phenotype.Citation12 This accession, Antwerpen-1 (An-1), also displayed enhanced susceptibility to several other pathogens. In a microarray comparison between large pools of susceptible and resistant F3 progeny between Col-0 and An-1, an over-representation of genes higher expressed in the resistant pool within a region at chromosome 4 was discovered. A similar genetic bias has previously been described for near isogenic lines of Ler-2 background carrying introgressions of Cvi-1.Citation15 Genetic mapping confirmed that this region indeed was responsible for the broad range resistance against several necrotrophic fungi, such as L. maculans, B. cinerea, Alternaria brassicicola and A. brassicae. This resistance trait was found to be unlinked to the two previously identified resistance loci in Col (RLM1) and Ler (RLM2), since F1 progeny between An-1 and a susceptible (rlm1rlm2) line of Col x Ler origin was found to be resistant against L. maculans and the new locus was thus named RLM3.Citation14 T-DNA insertion mutant screening of the differentially expressed genes identified one, At4g16990 (RLM3), as important for resistance. Northern blot analysis and RT-PCR detected several alternative transcripts of this gene. None of the T-DNA mutants showed as severe phenotypes as the naturally occurring deletion mutation found in the An-1 accession, but none of the 5 allelic T-DNA mutants in Col-0 background was a complete knockout. However, during complementation studies by overexpression of the shortest identified transcript—many T-DNA mutant plants showed increased susceptibility. RT-PCR showed that the increased susceptibility in some lines was due to co-suppression and silencing of the remaining RLM3 transcripts, giving further support of its role in general resistance towards several necrotrophic fungi. Some true overexpressors also showed a reduced susceptibility. Complementation by a genomic clone of RLM3 resulted in complete restoration of resistance in An-1, which indicate that multiple transcripts of RLM3 may be important.
Truncated NB-LRR Encoding Genes and Alternative Transcripts
The Arabidopsis Col-0 genome contains 83 TIR-NB-LRR, and 51 CC-NB-LRR encoding genes together with a number of truncated relatives (4 CC-NB, 21 TIR-NB, 1 CC-X, 30 TIR-X, 6 NB-LRR and 11 with complex domain compositions) of these two major R gene families.Citation16 It was early suggested that the truncated forms may act as adaptors for downstream signaling events analogously to the role of MyD88 downstream of most toll-like receptors (TLRs) in animal innate immunity.Citation17 Such adaptors should then be responsible for a broader resistance than each specialized receptor. The first truncated R gene-like gene to be reported as involved in disease resistance was RPW8, a member of the CC-X family, did indeed regulate a broad-range resistance against the powdery mildew pathogens Erysiphe spp.Citation18 and overexpression leads to a general increase in basal resistance towards biotrophs.Citation19 RLM3 has been annotated as part of the TIR-NB family of truncated R genes, but the shortest alternative transcript observed contained an alternative splicing of exon 2, encoding the NB-ARC/NOD domain, which made RLM3 encode a TIR-X transcript.Citation14 Alternative splicing has been demonstrated to be important for R gene function via the Pseudomonas syringae resistance gene RPS4.Citation20,Citation21 Interestingly, the different splice forms showed different protein stability, and it is anticipated that the RPS4 function is regulated at multiple levels to fine-tune its activity. The exact role and importance of the different RLM3 transcripts is currently not known and requires further evaluation.
The RCC1-like Domain of RLM3—an Adaptor with Functional Conservation Across Phyla?
An alternative transcript of RLM3 shows a 74 amino acid stretch encoded by the end of exon 1 and the exons 3 and 4 with sequence similarity to a section of amino acids close to the C terminus of the regulator of chromatin condensation (RCC1) family of proteins. This segment of amino acids contains the plant-specific DZC (disease resistance, zinc finger, chromosome condensation, IPR013591) domain, where At4g16990 (RLM3) is the only representative of the R gene family in Arabidopsis according to the PFAM and TAIR database protein domain search (). In rice, a CC-NB-LRR like protein (A2Y6C5, closest Arabidopsis homolog: RPP13) also carries the same domain, indicating that this motif unit may indeed be functionally or evolutionary relevant in certain types of disease resistance. This domain has also been named the BRX domain (brevis radix) and has been shown to mediate homo- and heterotypic protein-protein interactions.Citation22 Several distinct domains are common between plant and metazoan immunity indicating that a core-set of domains have been involved in this function already since the ancient eukaryotes.Citation23 Until recently, data on innate immunity outside the plant and metazoan phyla has been scarce. Interestingly, the recently identified protist slime mould protein TirA is an RCC1-like protein with a TIR domain.Citation24 TirA is required for specialized immune cell (called sentry cells, or S-cells) function in slime mould in its multi-cellular slug-form. Since this organism diverged from the metazoan/fungal lineage just after the split between the ancestor species that gave rise to plants and metazoans/fungi, it implies that the link to the RCC1 family of proteins is yet another aspect of a conserved core component in innate immunity from the ancient ancestor of all crown group phyla. The combination of two protein interaction domains may suggest that RLM3 indeed acts as a TIR-DZC adaptor between specific TIR-NB-LRR receptors and downstream components harboring the DZC domain. This downstream component could, among other things, be FYVE zinc finger proteins binding phosphoinositide at the membrane, like PARF-1,Citation25 or nuclear RCC1 proteins. The BRX family has also been suggested to act as transcription factors,Citation26 which could indicate a direct transcriptional regulation from RLM3 via this family. One possible target gene of such direct regulation (repression) would be WRKY60, which was the only gene consistently higher expressed in rlm3 compared to RLM3—material.Citation14 A future challenge will be to confirm and characterize the role of the DZC domain in plant immunity to fungal necrotrophs by dominant-negative RLM3 DZC domain overexpression constructs in the nucleus, cytoplasm or other cellular compartments and characterization of protein interaction targets. Alternatively, this might be accomplished by chimeric ubiquitin ligases degrading RLM3 DZC domain-interacting proteins.Citation27
Defense Responses Regulated by RLM3
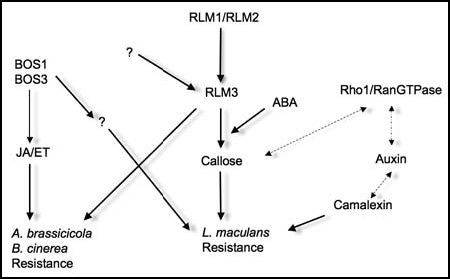
Assessments of the rlm3 genotypes for known defense responses against L. maculans and other pathogens did not reveal any effect on camalexin induction or JA/ET dependent transcription. This was surprising, since these are classical markers of a mounted defense against necrotrophs. It was also shown that RLM3 acts independently of BOS1 and BOS3, which both cause susceptible response towards both L. maculans and B. cinereaCitation14 (). On the other hand, callose induction was clearly impaired analogously to what had previously been seen on L. maculans responses of Arabidopsis with rlm1rlm2 genotype. This in combination with genetic evidence leads to the conclusion that RLM3 likely acts downstream of RLM1/RLM2 to mount certain defenses against L. maculans. Interestingly, the rlm3 genotypes displayed a slightly decreased susceptibility towards P. syringae. An intriguing possibility would be that RLM3 acts as a signaling switch between callose and SA-dependent responses, since P. syringae is known to influence this mutually antagonistic response via RIN4 in order to promote infection.Citation28 The enhanced susceptibility of RPW8 overexpressing plants when challenged by necrotrophic fungiCitation19 could however also indicate that RLM3 and RPW8 regulate basal responses that are mutually antagonistic—leading to a general role of short R gene -like proteins in early recognition events and signaling. This may suggest that the absence of RLM3 influences the transcriptional responses and the defense outcome also in other systems than the necrotrophic fungi.
Abbreviations
| RLM3 | = | resistance to Leptosphaeria maculans 3 |
| TIR | = | toll- and interleukin-1 receptor domain |
| CC | = | coiled-coil domain |
| NB | = | nucleotide binding domain (synonymous names: NB-ARC, NOD) |
| LRR | = | leucine rich repeat |
Figures and Tables
Figure 1 DZC containing proteins and their domain organization in Arabidopsis. In Arabidopsis, three main protein structure organizations containing the DZC domain are found. Structural diverse N-terminal domains include TIR, NOD and PH, RLM3 is the unique representative of the disease resistance family containing DZC at the C terminus within Arabidopsis. The BRX group is required for optimal root growth and RCC regulate chromatin condensation.
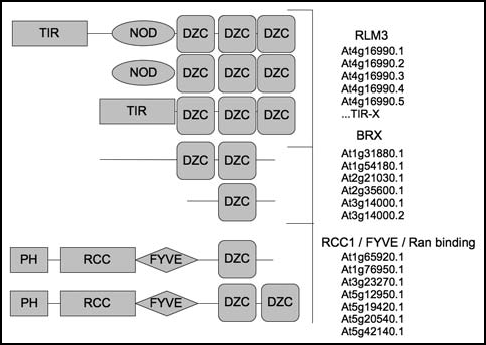
Figure 2 Overview of the parallel resistance pathways contributing to necrotroph resistance. RLM3 is proposed to act downstream of RLM1 or RLM2 in L. maculans resistance. However, genotypes lacking RLM1 and RLM2 are still resistant towards B. cinerea and A. brassicicola, indicating that other currently unknown upstream receptors for these pathogens remains to be found. On the other hand, the BOS1 and BOS3 genes are required for both L. maculans and other necrotrophic fungi, but these are induced independently of RLM3. In addition, auxin signaling influence L. maculans resistanceCitation29 but hitherto in an unknown manner (dotted arrows).
References
- Tauger M. Entitlement, shortage and the 1943 Bengal Famine: Another look. J Peasant Studies 2003; 31:45 - 72
- Maor R, Shirasu K. The arms race continues: battle strategies between plants and fungal pathogens. Curr Opin Microbiol 2005; 8:399 - 404
- OECD-FAO Agricultural outlook 2007–2016. 2007; OECD publishing ISBN: 9789264025097.
- Alfano J, Collmer A. Bacterial pathogens in plants: life up against the wall. Plant Cell 1996; 8:1683 - 1698
- Denby K, Kumar P, Kliebenstein D. Identification of Botrytis cinerea susceptibility loci in Arabidopsis thaliana. Plant J 2004; 38:473 - 486
- Glazebrook J. Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annu Rev Phytopathol 2005; 43:205 - 227
- Oliver R, Ipcho S. Arabidopsis pathology breathes life into the necrotroph-vs-biotrophs classification of fungal pathogens. Mol Plant Pathol 2004; 5:347 - 352
- Fitt B, Brun H, Barbetti M, Rimmer S. World-wide importance of phoma stem canker (Leptosphaeria maculans and L. biglobosa) on oilseed rape (Brassica napus). Eur J Plant Pathol 2006; 114:3 - 15
- Staal J, Kaliff M, Bohman S, Dixelius C. Transgressive segregation reveals two Arabidopsis TIR-NB-LRR resistance genes effective against Leptosphaeria maculans, causal agent of blackleg disease. Plant J 2006; 46:218 - 230
- Saal B, Struss D. RGA- and RAPD-derived SCAR markers for Brassica B-genome introgression conferring resistance to blackleg in oilseed rape. Theor Appl Genet 2005; 111:281 - 290
- Elliot C, Harjono Howlett B. Mutation of a gene in the fungus Leptosphaeria maculans allows increased frequency of penetration of stomatal apertures of Arabidopsis thaliana. Mol Plant 2008; E-pub.
- Bohman S, Staal J, Thomma BPHJ, Wang M, Dixelius C. Characterisation of an Arabidopsis—Leptosphaeria maculans pathosystem; resistance partially requires camalexin biosynthesis and is independent of salicylic acid, ethylene and jasmonic acid signalling. Plant J 2004; 37:9 - 20
- Kaliff M, Staal J, Myrenås M, Dixelius C. ABA is required for Leptosphaeria maculans resistance via ABI1 and ABI4 dependent signaling. Mol Plant-Microbe Interact 2007; 20:335 - 345
- Staal J, Kaliff M, Dewaele E, Persson M, Dixelius C. RLM3, a TIR domain-encoding gene involved in broad-range immunity of Arabidopsis to necrotrophic fungal pathogens. Plant J 2008; E-pub.
- Juenger T, Wayne T, Boles S, Symonds V, McKay J, Coughlan S. Natural genetic variation in whole-genome expression in Arabidopsis thaliana: the impact of physiological QTL introgression. Mol Ecol 2006; 15:1351 - 1365
- Meyers BC, Kozik A, Griego A, Kuang H, Michelmore RW. Genome-wide analysis of NB-LRR-encoding genes in Arabidopsis. Plant Cell 2003; 15:809 - 834
- Meyers BC, Morgante M, Michelmore RW. TIR-X and TIR-NB proteins: two new families related to disease resistance TIR-NB-LRR proteins encoded in Arabidopsis and other plant genomes. Plant J 2002; 32:77 - 92
- Xiao S, Ellwood S, Calis O, Patrick E, Li T, Coleman M, Turner J. Broad-spectrum mildew resistance in Arabidopsis thaliana mediated by RPW8. Science 2001; 291:118 - 120
- Wang W, Devoto A, Turner J, Xiao S. Expression of the membrane-associated resistance protein RPW8 enhances basal defense against biotrophic pathogens. Mol Plant Microbe Interact 2007; 20:966 - 976
- Zhang XC, Gassmann W. RPS4-mediated disease resistance requires the combined presence of RPS4 transcripts with full-length and truncated open reading frames. Plant Cell 2003; 15:2333 - 2342
- Zhang XC, Gassmann W. Alternative splicing and mRNA levels of the disease resistance gene RPS4 are induced during defense responses. Plant Physiol 2007; 145:1577 - 1587
- Briggs G, Mouchel C, Hardtke C. Characterization of the plant-specific BREVIS RADIX gene family reveals limited genetic redundancy despite high sequence conservation. Plant Physiol 2006; 140:1306 - 1316
- Staal J, Dixelius C. Tracing the ancient origins of plant innate immunity. Trends Plant Sci 2007; 12:334 - 342
- Chen G, Zhuchenko O, Kuspa A. Immune-like phagocyte activity in the social amoeba. Science 2007; 317:678 - 681
- Heras B, Dröbak B. PARF-1: an Arabidopsis thaliana FYVE-domain protein displaying a novel eukaryotic domain structure and phosphoinositide affinity. J Exp Bot 2002; 53:565 - 567
- Mouchel C, Briggs G, Hardtke C. Natural genetic variation in Arabidopsis identifies BREVIS RADIX, a novel regulator of cell proliferation and elongation in the root. Genes Dev 2004; 18:700 - 714
- Zhou P, Bogacki R, McReynolds L, Howley P. Harnessing the ubiquitination machinery to target the degradation of specific cellular proteins. Mol Cell 2000; 6:751 - 756
- Kim M, da Cunha L, McFall A, Belkhadir Y, DebRoy S, Dangl J, Mackey D. Two Pseudomonas syringae type III effectors inhibit RIN4-regulated basal defense in Arabidopsis. Cell 2005; 121:749 - 759
- Kaliff M. Genes, hormones and signalling pathways implicated in plant defence to Leptosphaeria maculans. Doctoral diss. Dept. of Plant Biology and Forest Genetics, SLU. Acta Universitatis agriculturae Sueciae 2007; 119