Abstract
The sea hare Aplysia is a powerful model organism for studying the structure and function of the nervous system. Recently, the genomic characterization of Aplysia has been facilitated: A large scale EST sequences was acquired by sequencing cDNA libraries from A. californica and a parallel EST database of the closely related species A. kurodai was reported. These EST databases provide useful tools for both molecular biology and bioinformatics. In our previous report, we demonstrated the utility of the database by screening the candidate genes for the synaptic plasticity and the behavioral sensitization using the microarray containing A. kurodai ESTs. In this Addendum, we have expanded our study to show that the protein domain repertoire and the abundance of regulatory genes displayed a linear relationship with the evolution of the complex brains in different lineages. This distinct set of protein domains may play critical roles in evolution of the nervous systems.
The marine mollusk Aplysia is an important model organism for studying the cellular and molecular mechanisms underlying learning and memory.Citation1–Citation3 To facilitate the molecular level analyses in Aplysia, a large scale of expressed sequence tag (EST) analyses has been performed in Aplysia californica.Citation4 Recently, we analyzed the ESTs from A. kurodai, another commonly used species of Aplysia in the laboratories in Northwest Asian Pacific area.Citation5 In this study, we collected 11,493 ESTs from the central nervous system of A. kurodai, which were assembled into 4,859 tentative transcripts. Sixty seven percent (3,267 contigs) of the 4,859 A. kurodai contigs were matched to known A. californica sequences and 1,592 (33%) had no significant matches (BLASTN; E ≤ 10−10) (all the sequence information is available at www.seahare.org). This additional EST database will facilitate the assembly, annotation of the forthcoming Aplysia genomic sequences. Moreover, using microarray we characterized the Ap-eIF3e as a positive regulator which is involved in the consolidation of long-term facilitation. The overexpression of Ap-eIF3e lowered the threshold for the long-term facilitation, whereas the inhibition of Ap-eIF3e impaired the long-term facilitation.Citation5
In this addendum, we investigated whether a set of nervous system-related genes have a correlation with the evolution of the organism's complexity and nervous systems across phyla by studying the relationship between distantly related organisms and their protein domains.Citation6 To this end, 184 and 134 Pfam (www.sanger.ac.uk/Software/Pfam/) protein domains were identified from the selected nervous system-related genes and housekeeping genes that previously collected by Dorus et al.Citation7 (available from www.seahare.org). GO analysis showed 76.5% of the protein domains from the selected housekeeping genes are involved in basic metabolism such as protein biosynthesis and electron transport, whereas the diverse functions in signal transductions were assigned to the domains from the selected nervous system-related genes (data not shown).
First, we analyzed the repertoires of those protein domains in seventeen organisms (Aplysia californica, Saccharomyces cerevisiae, Dictyostelium discoideum, Caenorhabditis elegans, Anophelies gambiae, Drosophila melanogaster, Ciona intestinalis, Danio rerio, Xenopus tropicalis, Gallus gallus, Monodelphis domestica, Bos taurus, Canis familiaris, Rattus norvegicus, Mus musculus, Pan trogiodytes, Homo sapiens). The ratio (relative abundance) of domain repertoires in signal transduction-related genes to those in the housekeeping metabolic genes was used as an index. Interestingly, among the seventeen organisms, two non-animal species, Dictyostelium and Saccharomyces, showed the lowest ratios (below 1.0), while all the other animals showed ratios >1.0 (). Vertebrates showed significantly higher ratios of domain repertoires (nervous system-related versus housekeeping) than selected invertebrates (1.26 ± 0.02 vs. 1.38 ± 0.00, p < 0.0001), suggesting that the type of protein domain families has increased in the vertebrate lineage (). Additionally, domain repertoires of mammals were not significantly different from those of selected non-mammalian vertebrates confirming parallel evolution in major vertebrate classes. Next, we calculated the total domain abundance, which is the number of proteins in an organism that contain domains related to signaling or domains involved in basic metabolism (). The domain abundance ratio of signaling to metabolic (or housekeeping) genes was significantly higher in the vertebrates than in the majority of invertebrate species (2.78 ± 0.08 vs. 2.13 ± 0.14, respectively, p < 0.01) (). However, C. elegans showed an exceptionally high abundance ratio compared to other invertebrates selected here (). C. elegans has a comparable diversity of signal transduction-related domains to those of several vertebrate animals. This might be related to a distinct adaptive strategies in this nematode lineage and the fact that C. elegans has many protein domains that interact with the environment using extracellular chemical ligands as signals.Citation8
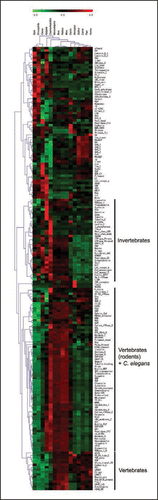
To identify the specific protein domains whose expansion may have contributed to the evolution of the signaling mechanisms associated to nervous system of selected 15 species, we performed a clustering analysis using the abundance of the 184 signal transduction or cell-cell interactions-related protein domains (). As expected, clustering results revealed that different organisms are enriched with different set of domains further supporting different evolutionary strategies in each animal group.
For example, in Aplysia, Sema domain was found to be the most highly expanded. Sema domain, which constitutes the distinctive structural and functional element of semaphorins, is known for its roles in axonal pathfinding during development, tissue differentiation and regeneration.Citation9,Citation10 It was surprising to find that this domain expanded more in Aplysia than vertebrates since nineteen semaphorines have been reported in vertebrates, while only three in invertebrates such as Drosophila.Citation9 However, even in vertebrates, the biological functions of many semaphorine families remain to be elucidated. Taken together, we suggest that the domain clusters specifically enriched in a certain organism, such as Sema domain in Aplysia, might have played important roles in adaptive strategies associated with speciation in this gastropod lineage.
Figures and Tables
Figure 1 Comparative analysis of the neural signaling related protein domains. (A1 and B1) The ratio of domain repertoire (signaling/housekeeping) (A1) and domain abundance (B1) in 17 organisms from unicellular eukaryotes to mammals. (A2 and B2) The averaged ratio values from invertebrates, non-mammalian vertebrates, mammals and other organisms were plotted (mean ± SEM). The connecting lines are only for a better visualization.
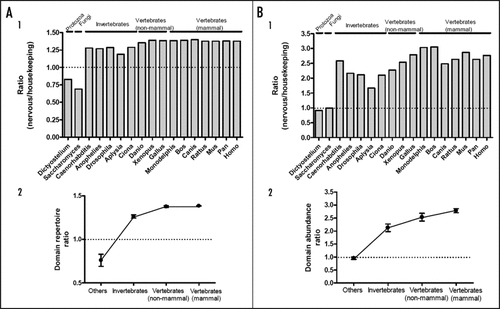
Figure 2 Hierarchical clustering of the abundance of the neural signaling related domains. Each column and row represents one species and protein domain, respectively. The abundance of individual domains was normalized both within and across different species: red, high relative abundance, green, low relative abundance. Representative large clusters are indicated.
Acknowledgements
This work was supported by National Creative Research Initiative Program of the Korean Ministry of Science and Technology and the Marine and Extreme Genome Research Center Program, Ministry of Marine Affairs and Fisheries, Republic of Korea. KOBIC was supported by the Korean Ministry of Science and Technology (MOST) under grant number M10407010001-04N0701-00110 and by the MIC (Ministry of Information and Communication), Korea, under the KADO (Korea Agency Digital Opportunity & Promotion) support program (06-121).
Addendum to:
References
- Bailey CH, Bartsch D, Kandel ER. Toward a molecular definition of long-term memory storage. Proc Natl Acad Sci USA 1996; 93:13445 - 13452
- Carew TJ, Sahley CL. Invertebrate learning and memory: from behavior to molecules. Annu Rev Neurosci 1986; 9:435 - 487
- Kandel ER. The molecular biology of memory storage: a dialogue between genes and synapses. Science 2001; 294:1030 - 1038
- Moroz LL, Edwards JR, Puthanveettil SV, Kohn AB, Ha T, Heyland A, et al. Neuronal transcriptome of aplysia: neuronal compartments and circuitry. Cell 2006; 127:1453 1467
- Lee YS, Choi SL, Kim TH, Lee JA, Kim HK, Kim H, et al. Transcriptome analysis and identification of regulators for long-term plasticity in Aplysia kurodai. Proc Natl Acad Sci USA 2008; 105:18602 - 18607
- Murzin AG, Brenner SE, Hubbard T, Chothia C. SCOP: a structural classification of proteins database for the investigation of sequences and structures. J Mol Biol 1995; 247:536 - 540
- Dorus S, Vallender EJ, Evans PD, Anderson JR, Gilbert SL, Mahowald M, et al. Accelerated evolution of nervous system genes in the origin of Homo sapiens. Cell 2004; 119:1027 - 1040
- Robertson HM. Two large families of chemoreceptor genes in the nematodes Caenorhabditis elegans and Caenorhabditis briggsae reveal extensive gene duplication, diversification, movement and intron loss. Genome Res 1998; 8:449 - 463
- Gherardi E, Love CA, Esnouf RM, Jones EY. The sema domain. Curr Opin Struct Biol 2004; 14:669 - 678
- Raper JA. Semaphorins and their receptors in vertebrates and invertebrates. Curr Opin Neurobiol 2000; 10:88 - 94