Abstract
Chemical signaling plays a major role in shaping life history processes that drive ecology and evolution in marine systems, notably including habitat selection by marine invertebrate larvae that must settle out of the plankton onto the benthos.1 For larvae, the identification of appropriate habitats in which to settle and undergo metamorphosis to the adult form relies heavily on the recognition of cues indicative of a favorable environment. By documenting settlement responses of larvae of the tropical abalone, Haliotis asinina, to a range of coralline algae species, we recently highlighted the species-specific nature of this interaction.2 Here, we demonstrate that this specificity is likely driven by chemical, rather than physical, properties of the algae. Our initial characterization of the surface cell biomarkers from three different algal species show that inductive cue biomolecular composition correlates with variations in larval settlement response.
Studies on several abalone (Gastropoda, Haliotidae) species indicate that larvae can distinguish between different coralline algae species and identify preferred species/sites for settlement.Citation3,Citation4 Each abalone species surveyed to date is induced most effectively by a different species of coralline alga.Citation5 Recently, we have documented the induction response of larvae of the tropical abalone, Haliotis asinina, to a range of coralline algae species collected from Heron Island Reef, Australia (23°27′S; 151°55′E), a natural habitat of adult H. asinina.Citation2 We found that H. asinina larvae displayed a broad range of responses across the 16 coralline species tested, from 0–100% metamorphosis by 48 hours post induction (hpi). Unlike any abalone species previously studied, the greatest numbers of H. asinina were induced to metamorphose by densely branched articulated corallines of the genus Amphiroa (92.8–100% metamorphosis by 48 hpi) ().
Across the 16 algal species assayed, we observed a strong trend towards increased settlement response associated with increased branching morphology,Citation2 suggesting that physical cues (e.g., algal morphology and texture), rather than chemical cues, may be the primary factor influencing larval settlement response. To distinguish between these two alternatives we repeated larval settlement assays as described in,Citation2 this time comparing the response of larvae exposed to three species of coralline algae with different morphologies, presented as either live shards or dead, air dried shards. Coralline species used were the encrusting Hydrolithon onkodes, the branching Lithophyllum moluccense and the articulated Amphiroa ephedraea, previously shownCitation2 to induce poor, moderate and high levels of settlement, respectively, when presented as live shards. Two-way ANOVA (p-value < 0.05) indicates that larval settlement response at 12 hpi varies significantly between coralline species when the algae are alive, but not when dead and air-dried (). Furthermore, larval settlement overall at 12 hpi was significantly greater on live coralline pieces compared to dead ones. We therefore conclude that the species-specific response shown by H. asinina larvae induced by different coralline species is the result of interactions between the larvae and chemical (not physical) cues provided by the algae.
To date, gamma aminobutyric acid (GABA), delta-aminovaleric acid (5-AVA), L-glutamic acid (GA), monosodium glutamate (MSG), dibromomethane, hydrogen peroxide and aspartic acid all have been found to induce low to moderate levels of larval settlement, metamorphosis, or both, in one or more abalone species, highlighting the complexity of this process.Citation6–Citation11 Low molecular weight protein-associated molecules extracted directly from cyanobacteria or coralline algae have also been used successfully to induce settlement and metamorphosis in two abalone species.Citation12,Citation13 Although the chemical nature of these molecules remains elusive, they may provide the most ecologically relevant clues for abalone larval induction, as various coralline algae have proven to be the most effective known inducers of metamorphosis among multiple abalone species. We chose to characterize low molecular weight masses (biomarkers) of surface cells isolated from three coralline algae species capable of inducing metamorphosis in Haliotis asinina—a poor, an intermediate and an excellent inducer. Our previous data showed that by 48 hpi, Hydrolithon onkodes induced a mean of 31.3% metamorphosis, while Lithophyllum moluccense induced a mean of 77.6% metamorphosis and Amphiroa ephedraea induced a mean of 95.4% metamorphosis.Citation2
Surface cell biomarkers were characterized in the three coralline species using C18 extraction and matrix-assisted laser desorpt-ionionization-time of flight mass spectrometry (MALDI-TOF MS). Mass spectrometry results reveal that each coralline species has its own unique surface cell chemical profile (). MALDI-TOF MS has not previously been employed to distinguish between coralline algae species, however it has proven useful in the identification of bacteria, fungi, viruses and dinoflagellates.Citation14,Citation15 Our results indicate that it may be possible to employ similar methods for coralline species identification, including for taxonomic purposes. Overall, Amphiroa ephedraea and Lithophyllum moluccense had more similar surface cell biomarker profiles compared to Hydrolithon onkodes (). This is reflective of the phylogenetic relationship of the three species. Molecular studies show that A. ephedraea and L. moluccense belong to the same family, Lithophylloideae, while H. onkodes is a member of the family Mastophoroideae.Citation16,Citation17 MALDI-TOF MS analysis returned no significant peak signals in the 1,000–2,000 mass/charge (m/z) range.
Molecules at 690 and 890 daltons (Da) isolated from the coralline alga Lithothamnium californicum induced high levels of metamorphosis in the red abalone, Haliotis rufescens.Citation12 Similarly, 600–1,100 Da extracts of the coralline alga Phymatolithon repandum induced metamorphosis in Haliotis virginea.Citation18 Our mass spectrometry analysis has identified biomarkers between 441 and 928 Da in the corallines Hydrolithon onkodes, Lithophyllum moluccense and Amphiroa ephedraea and it is likely that at least some of these molecules play a role in chemical signaling during the induction of H. asinina metamorphosis. Mass spectrometry analysis of three coralline species reveals that two biomarkers—one 620.8 Da and one 741.7 Da—are present in the surface cells of all three species. Overall, A. ephedraea and L. moluccense had more similar profiles, with five mass peaks in common, plus four and two unique peaks respectively. In contrast, H. onkodes has five unique peaks. Interestingly, A. ephedraea and L. moluccense are much more effective inducers of Haliotis asinina metamorphosisCitation2 than is H. onkodes, and this may be related to the similarities found here in their surface cell biomarker profiles. Isolation of biomolecules playing a primary role in chemical signaling between larvae and algae during settlement and metamorphosis will further our understanding of how this crucial interaction has influenced the ecology and evolution of H. asinina.
C18 ZipTip Purification of Algal Extracts and MALDI-TOF MS Methods
Five separate plants of each of the three coralline algae species were collected from Heron Island Reef. Within 1 h of collection, ∼5 µl surface cells were scraped from each plant/colony using a scalpel. Algal surface cells were resuspended in 100 µl 0.1% trifluoroacetic acid (TFA), mixed by vortexing and rotated on a wheel for 15 min. Inorganic salts in the samples were removed by absorbing 20 µl sample with 0.1% TFA solution onto C18 ZipTips (Millipore, MA). ZipTips were washed five times with 10 µl 0.1% TFA/5% MeOH. Biomolecules were eluted from zip tips with 10 µl 0.1% TFA/50% acetonitrile (CH3CN). 0.5 µl of eluted sample was mixed with 0.5 µl of matrix solution (20 mg/ml 2,5-dihydrobenzoic acid (DHB) in 30% CH3CN/0.1% TFA) and spotted onto a mass spectrometer target plate (Applied Biosystems USA, P/N:V700666). Each of the five biological replicates for each of the three coralline algae species were spotted on to the plate five times, in addition to five replicates of 1 µl matrix solution as a control (80 spots total). After drying at ambient temperature, samples were analyzed by MALDI-TOF MS performed on a Voyager-DE STR Biospectrometry workstation (PerSeptive Biosystems) equipped with a N2 laser and pulsed ion extraction accessory. The instrument was calibrated using ProteoMass Peptide MALDI-MS calibration standards (Sigma). Final spectra resulted from 500 shots, recorded in the reflectron mode within a mass range from m/z 420 to m/z 1,000 and 1,000 to 2,000 Da.
Data were progressively condensed such that any mass peaks that occurred in less than three of the five technical replicates, and in less than three of the five biological replicates, for each algal species were removed from further analysis. The condensed profile for each algae was subtracted from matrix controls. Finally, algal profiles were compared to each other to establish similarities and differences in surface cell biomarker composition.
Abbreviations
| hpi | = | hours post induction |
| ANOVA | = | analysis of variance |
| SEM | = | scanning electron microscopy |
| MALDI-TOF MS | = | matrix-assisted laser desorption-ionization-time of flight mass spectrometry |
| TFA | = | trifluoroacetic acid |
| DHB | = | 2,5-dihydrobenzoic acid |
| ACN | = | acetonitrile |
| C18 | = | carbon isotope 18 |
| GABA | = | gamma aminobutyric acid |
| 5-AVA | = | delta-aminovaleric acid |
| GA | = | L-glutamic acid |
| MSG | = | monosodium glutamate |
| Da | = | daltons |
| m/z | = | mass-to-charge ratio |
Figures and Tables
Figure 1 A newly metamorphosed Haliotis asinina post-larvae explores the surface of Amphiroa ephedraea, its coralline inductive cue. Amphiroa spp. are the most effective natural inducers of H. asinina settlement and metamorphosis.Citation2 SEM photograph at 110x magnification. Photo credit: Erica Lovas.
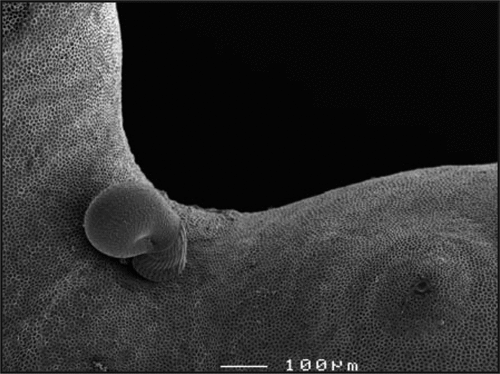
Figure 2 Results of a settlement assay comparing the effectiveness of dead and live coralline algae as inducers of Haliotis asinina metamorphosis. 2-way ANOVA (p-value < 0.05) indicates that live coralline algae induced significantly higher numbers of larval settlement than dead coralline algae by 12 hpi, suggesting that H. asinina induction is mediated by chemical, not physical cues. Numbers of larvae settled on live and dead algae of the encrusting Hydrolithon onkodes (H.o), the branching Lithophyllum moluccense (L.m) and the articulated Amphiroa ephedraea (A.e) were counted at 12 hpi.
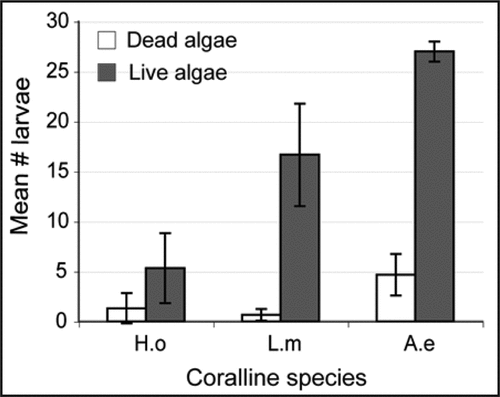
Table 1 Surface cell biomarker profiles, as indicated by MALDI-TOF mass spectrometry analysis, of three species of coralline algae capable of inducing Haliotis asinina settlement and metamorphosis
Acknowledgements
This work was funded by an Australian Research Council (ARC) grant to Sandie M. Degnan.
Addendum to:
References
- Zimmer RK, Butman CA. Chemical signaling processes in the marine environment. Biol Bull 2000; 198:168 - 187
- Williams EA, Craigie A, Yeates A, Degnan SM. Articulated coralline algae of the genus Amphiroa are highly effective natural inducers of settlement in the tropical abalone, Haliotis asinina. Biol Bull 2008; 215:98 - 107
- Daume S, Brand-Gardner S, Woelkerling WJ. Settlement of abalone larvae (Haliotis laevigata Donovan) in response to non-geniculate coralline red algae (Corallinales, Rhodophyta). J Exp Mar Biol Ecol 1999; 234:125 - 143
- Roberts RD, Kaspar HF, Barker RJ. Settlement of abalone (Haliotis iris) larvae in response to five species of coralline algae. J Shellfish Res 2004; 23:975 - 987
- Roberts RD. A review of settlement cues for larval abalone (Haliotis spp.). J Shellfish Res 2001; 20:571 - 586
- Morse DE, Hooker N, Duncan H, Jensen L. g-aminobutyric acid, a neurotransmitter, induces planktonic abalone larvae to settle and begin metamorphosis. Science 1979; 204:407 - 410
- Seki T, Taniguchi K, Kurata K. The metamorphosis inducing role of dibromomethane on the Japanese abalone, Haliotis discus hannai Ino 1997; Abstracts of the Third International Abalone Symposium Monterey 60
- Suenaga K, Hoki H, Ishida H, Nukaya H, Roberts RD, Tsuji K. Inducing substance for abalone metamorphosis from the crustose coralline algae Hydrolithon samoense. Fisheries Sci 2004; 70:342 - 344
- Gordon N, Shpigel M, Harpaz S, Lee JJ, Neori A. The settlement of abalone (Haliotis discus hannai Ino) larvae on culture layers of different diatoms. J Shellfish Res 2004; 23:561 - 568
- Zhang XJ, Yang ZH, Cai ZH. The hydrogen peroxide impact on larval settlement and metamorphosis of abalone Haliotis diversicolor supertexta. Chin J Oceanol Limnol 2008; 26:238 - 241
- Stewart P, Soonklang N, Stewart MJ, Wanichanon C, Hanna PJ, Poomtong T, et al. Larval settlement of the tropical abalone, Haliotis asinina Linnaeus, using natural and artificial chemical inducers. Aquac Res 2008; 39:1181 - 1189
- Morse ANC, Froyd CA, Morse DE. Molecules from cyanobacteria and red algae that induce larval settlement and metamorphosis in the mollusc Haliotis rufescens. Mar Biol 1984; 293 - 298
- Roberts RD, Nicholson CM. Variable response from abalone larvae (Haliots iris, H. virginea) to a range of settlement cues. Moll Res 1997; 18:131 - 141
- Fenselau C, Demirev PA. Characterization of intact microorganisms by MALDI mass spectrometry. Mass Spectrom Rev 2001; 20:157 - 171
- Lee FW, Ho KC, Lo SC. Rapid identification of dinoflagellates using protein profiling with matrix-assisted laser desorption/ionisation mass spectrometry. Harmful Algae 2008; 7:551 - 559
- Bailey JC, Gabel JE, Freshwater DW. Nuclear 18S rRNA gene sequence analyses indicate that the Mastophoroideae (Corallinaceae, Rhodophyta) is a polyphyletic taxon. Phycologia 2004; 43:3 - 12
- Broom JES, Hart DR, Farr TJ, Nelson WA, Neill KF, Harvey AS, et al. Utillity of psbA and nSSU for phylogenetic reconstruction in the Corallinales based on New Zealand taxa. Mol Phylogenet Evol 2008; 46:958 - 973
- Roberts RD, Kaspar H, Mountfort D, Berkett N. Induction of settlement and metamorphosis in paua (Haliotis spp.) by algae and algal extracts 1994; Abstracts of the New Zealand Marine Sciences Society Annual Conference Hamilton 61