Abstract
Specific questions about intron evolution are precisely addressed applying a phylogenomic approach to suitable gene families. With this approach we have recently reported that the appearance of most human tetraspanins occurred in the common ancestor of vertebrates and coincides in nearly all cases with the concomitant acquisition of new introns. We observed that indels at the ends of the DNA exonic sequences with no involvement of the corresponding intronic sequence, were the cause of two discordant intron positions between orthologous tetraspanins. Here, we discuss a putative intron sliding occurrence in which a new acquired intron junction (intron 1a) in the ancestor of chordates could have been shifted to new positions (introns 1b and 1c) during the expansion of the tetraspanin family in vertebrates. Such a mechanism could be responsible for generating some of the variation of function in this important family of membrane spanning proteins.
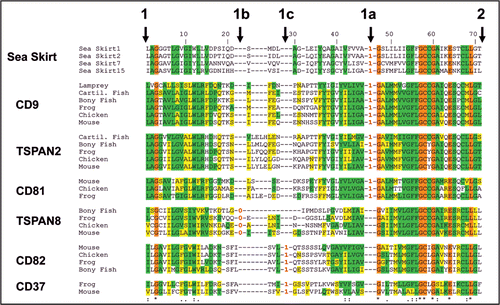
Intron sliding (IS) is defined as a relocation or shift of an intron to a different position of a gene over a short distance of base pairs. Although intron sliding has been suggested as highly likely in certain situations, IS has been considered an infrequent phenomenon, since imperfect protein alignments can be used to explain most of the observed putative IS occurrences. In addition, it is thought that the sliding event needs two simultaneous mutations to occur.Citation1–Citation3 Recently a new mechanism of IS formation has been proposed that does not rely on simultaneous mutation events and could explain the frequently observed alignment gaps near exon-intron junctions as well as discordant intron positions over short distances in homologous proteins.Citation4 This new mechanisms may explain the short deletions (indels) we observed near the intron-exon junctions in two cases of discordant intron positions in orthologs of the CD81 and TSPAN14 tetraspanins.Citation5 In the cases we reported, we compared highly conserved intron DNA sequences from closely related orthologs. We ruled out exonization or intronization (the assimilation of adjacent intronic sequences into an existing exon or exonic sequences into a existing intron) as a factor in the discordant intron-exon junctions.Citation5 Additionally, this mechanisms of IS formationCitation4 could explain a puzzling intron discordant distribution of new introns 1a, 1b and 1c in two clusters of tetraspanin paralogs CD9/TSPAN2/CD81 and TSPAN8/CD82/CD37 ().
Tetraspanins (tetraspan or TM4SF) are broadly expressed integral membrane proteins with 33 paralogs in the human genome. Animal tetraspanins are classified in four large families;Citation6 the CD family of tetraspanins contains all CD tetraspanins except CD63 and has several well defined clusters such as the CD9/TSPAN2/CD81 cluster, which is of chordate origin (525 Mya;Citation7), and the TSPAN8/CD82/CD37 cluster that appeared later in the common ancestor of vertebrates (450 Mya;Citation6,Citation8).
Using a phylogenomic approach we were able to identify the temporal distribution of the origins of the 19 introns that have been conserved in the above mentioned 33 human tetraspanin paralogs.Citation5 Of these 19 introns, 9 are phase 0 ancestral introns we called 1, 2, 3, 4, 4a, 4b, 4c, 5 and 6, that appeared before the common ancestor of Bilateria (i.e., before the split of protostomes and deuterostomes), the remaining 10 introns are of more recent origin appearing in the deuterostome branch and, unlike the older introns, most of them are of phases 1 or 2 (introns can be located between or within codons: introns of phase 0 are located between codons, while introns of phases 1 or 2 are within codons and break the reading frame). Intron 1a is present in the CD9/TSPAN2/CD81 tetraspanins cluster between ancestral introns 1 and 2 (), and is the oldest human homolog of these 10 more recent deuterostome specific introns. This intron can be inferred to exist in the common ancestor of urochordates (Ciona intestinallis; ); intron 1b in TSPAN8 and intron 1c in CD82/CD37 tertraspanins appeared later in the tree of life in the common ancestor of vertebrates (5; ).
Tetraspanins from the nonvertebrate deuterostome Ciona (sea squirt) might be basal to multiple ortholog groups as a result of duplication events in the vertebrates. Groups such as vertebrate TSPAN8 and CD82/CD37 could be derived from these as our phylogenetic analysis revealed, although with low robustness at the base.Citation6 If the Ciona CD9/TSPAN2/CD81 tetraspanins, which have intron position 1a, are ancestors of TSPAN8/CD82/CD37 tetraspanins, then the appearance of introns 1b and 1c in vertebrates tetraspanins TSPAN8/CD82/CD37 can be explained by at least one or two events of intron loss and two events of intron gain. Since both intron loss and gain in vertebrates is a rare phenomenon,Citation1,Citation5 and these three or four events of intron gain/loss occurred in close paralogs which will make the process even more infrequent, the view of introns 1b and 1c being a relocation of intron 1a during the expansion of tetraspanins in vertebrates seems highly plausible.
Since the new mechanisms of IS proposedCitation4 allows for the presence of transcripts with premature termination codons during the process of IS, it allows for the change of the phase of an intron during an IS event. To fully ascertain the process of the tetraspanin intron 1a putative sliding, more species' tetraspanins need to be analyzed. Because genes, including tetraspanin genes,Citation9 are selectively lost in some species, some events that are used to reconstruct evolutionary history could be missed as a result of the sparse taxonomic sampling we used for these analyses. In addition, we cannot rule out the possibility of introns arising more than once in the same position of a gene.Citation10 However, a phylogenomic approach of this phenomenon in a suitable protein family such as the tetraspanins (provided a sufficient number of representative species are available) can be extremely useful in the study of IS occurrence.
Figures and Tables
Figure 1 Sequence alignment of tetraspanins from the groups CD9/TSPAN2/CD81, CD82/CD37 and TSPAN8, depicting possible intron sliding positions 1b and 1c. The alignment shows the full sequence between ancestral intron positions 1 and 2. Vertical bars indicate exon/intron junctios 1, 1a, 1b and 1c and 2. In the alignment the presence of an intron is shown within the amino acid sequences by a red number, which indicates the intron phase (0 is between codons, 1 is between the first and second position of a codon). Shade colours code: orange identical in all sequences; green identical and yellow similar to Sea skirt sequences. Mouse, Mus musculus; Chicken, Gallus gallus; Frog, Xenopus tropicalis; Bony fish, (zebrafish) Danio rerio; Cartilaginous fish, (shark) Callorhinchus mili; Lamprey, Petromyzon marinus; Sea skirt, Ciona intestinalis.
Addendum to:
References
- RRogozin IB, Sverdlov AV, Babenko VN, Koonin EV. Analysis of evolution of exonintron structure of eukaryotic genes. Brief Bioinform 2005; 6:118 - 134
- Fedorov A, Merican AF, Gilbert W. Large-scale comparison of intron positions among animal, plant and fungal genes. Proc Natl Acad Sci USA 2002; 99:16128 - 16133
- Stoltzfus A, Logsdon JM Jr, Palmer JD, Doolittle WF. Intron “sliding” and the diversity of intron positions. Proc Natl Acad Sci USA 1997; 94:10739 - 10744
- Tarrío R, Ayala FJ, Rodríguez-Trelles F. Alternative splicing: a missing piece in the puzzle of intron gain. Proc Natl Acad Sci USA 2008; 105:7223 - 7228
- Garcia-España A, Mares R, Sun TT, Desalle R. Intron evolution: testing hypotheses of intron evolution using the phylogenomics of tetraspanins. PLoS ONE 2009; 4:4680
- Garcia-España A, Chung PJ, Sarkar IN, Stiner E, Sun TT, Desalle R. Appearance of new tetraspanin genes during vertebrate evolution. Genomics 2008; 91:326 - 334
- Shu D, Morris SC, Zhang ZF, Liu JN, Han J, Chen L, et al. A new species of yunnanozoan with implications for deuterostome evolution. Science 2003; 299:1380 - 1384
- Hedges SB, Kumar S. Genomics. Vertebrate genomes compared. Science 2002; 297:1283 - 1285
- Garcia-España A, Chung PJ, Zhao X, Lee A, Pellicer A, Yu J, et al. Origin of the tetraspanin uroplakins and their co-evolution with associated proteins: implications for uroplakin structure and function. Mol Phylogenet Evol 2006; 41:355 - 367
- Dibb NJ, Newman AJ. Evidence that introns arose at proto-splice sites. EMBO J 1989; 8:2015 - 2021