Abstract
The green alga Volvox carteri is one of the simplest multicellular organisms. It consists of only two cell types, somatic and reproductive cells, making it a suitable model system for studying cell division, multicellularity, and cellular differentiation. Each of the ~2000-4000 cells of an adult, asexual organism arises through a sequence of symmetric and asymmetric cleavage divisions from a single, asexual reproductive cell. As in ontogenetic development of higher organisms, the fate of a Volvox blastomere (i.e., whether it undergoes division or differentiation) is determined by a complex balance of regulators. Retinoblastoma-related proteins (RBRs) seem to act as key regulators and hubs in cell cycle control and, therefore, have been investigated in detail in higher organisms. Recently, the identification and characterization of a gender-specific RBR in Volvox, RBR1, revealed a role for the retinoblastoma protein family in sexual development. RBRs are elements of a conserved signal-transduction pathway called the retinoblastoma (RB) tumor suppressor pathway. In addition to RBR1, other key components of this pathway are present in Volvox, demonstrating that the RB signal-transduction pathway is utilized by these simple green algae.
In animals, retinoblastoma-related proteins (RBRs) are tumor suppressors that act as key regulators of cell cycle entry. RBRs are involved not only in the regulation of cell division but also in tumor suppression, DNA repair, DNA damage checkpoint control, differentiation, cellular senescence and apoptosis.Citation1–Citation5 RBRs act as hubs for controlling the cell cycle by connecting the cell cycle clock with transcription machinery. In this way, banks of genes are controlled via the retinoblastoma (RB) tumor suppressor pathway.Citation1,Citation6–Citation11
Recently, a retinoblastoma-related protein, RBR1, has been cloned and characterized from the green alga Volvox carteri.Citation12 Volvox is one of the simplest multicellular organisms because it is composed of only two cell types. All cells of an asexual adult Volvox, i.e., ∼2,000–4,000 biflagellate somatic (body) cells and ∼16 asexual germ (reproductive) cells, arise through 11 or 12 rapid symmetrical and asymmetrical cleavage divisions from a single, asexual reproductive cell. In addition to asexual reproduction, Volvox is also capable of sexual reproduction. Due to these features and several others, Volvox is an ideal model system for the molecular analysis of basic principles of the developmental processes related to cell division, multicellularity and cellular differentiation.Citation13,Citation14
The RBR1 protein identified in Volvox has the same sequence features as other RB family proteins, particularly a pocket domain.Citation12 The characteristics of RBR1 suggest that it plays a role in cellular differentiation and cell cycle control during Volvox embryogenesis, similar to the role of RB proteins in higher organisms. RBR1 is not only expressed under developmental control but also shows cell type-specific expression, with the highest mRNA level detected in somatic cells of adult Volvox.Citation12 The presence of four splice variants and many potential cyclin-dependent kinase phosphorylation sites suggests that RBR1 is subject to control at the posttranscriptional and posttranslational levels.Citation12 The most unusual characteristic of RBR1 is that the gene is only present in females, mapping to the female mating-type locus. However, results with transgenic males expressing the female RBR1 gene suggest that a functionally related analog of RBR1 exists in males.Citation12
Volvox RBR1 is just one component of the RB signal-transduction pathway. There are probably several targets for RBR1, but frequently the function of an RBR depends on its interactions with the E2F family of DNA-binding transcription factors,Citation15,Citation16 which are bound by RBRs. However, in order to interact with RB family proteins, E2Fs need to attach to E2F dimerization partner 1 (DP1) subunits and to stably bind E2F promoter elements. Binding of an RBR to the E2F/DP1/DNA complex blocks the ability of E2F/DP1 to activate transcriptionCitation4,Citation15,Citation16 (). Both E2F (E2F1, Acc. No. FJ898368) and DP1 (Acc. No. FJ898367) have been identified in female V. carteri. They show high degrees of sequence similarity with E2Fs or DP1s from distantly related species ( and B).
RBR hyperphosphorylation is known to cause release of the RBR from the RBR/E2F/DP1 complex ( and C), and this release allows E2F/DP1 transcription factors to drive cell proliferation.Citation16 Analysis of the Volvox RBR1 sequence reveals 15 potential phosphorylation sites for hyperphosphorylation by cyclin-dependent kinases (CDKs).Citation12 A search of the V. carteri female genome (v1.0) database at the Department of Energy (DOE) Joint Genome Institute (JGI) (http://genomeportal.jgi-psf.org/Volca1/) reveals that about ten CDKs are present in the V. carteri genome. As the name suggests, CDKs are controlled by cyclins, and one particular cyclin, cyclin D, is known to be the regulatory subunit of a CDK/D-type cyclin holoenzyme. D-type cyclin subunits with conserved LxCxE motifs (where x represents any amino acid) enable the CDK/D-type cyclin holoenzyme to bind the LxCxE binding site of RBRsCitation17,Citation18 (). An LxCxE binding site was also identified in Volvox RBR1,Citation12 and a BLAST search revealed the existence of several D-type cyclins in the female Volvox genome (DOE JGI Volvox genome portal v1.0). Just as with other D-type cyclins, Volvox D-type cyclins contain conserved cyclin boxes, and four of them contain the expected LxCxE motif close to their N-termini (see LxCxE motifs in ).
Certainly, the RBR pathway is expected to be quite complicated, containing several other binding partners and regulatory proteins, but this analysis shows that the main proteins involved in the RB pathway in higher organisms are present in female Volvox. Moreover, results with Volvox RBR1 demonstrate that the RB pathway is functional in these algae. However, the fact that males do not possess RBR1, although they likely possess a functionally related analog of RBR1, shows that there are gender-specific differences in the Volvox RB pathway. So far, there is little information about male Volvox, and there is almost no sequence information from the male genome. Therefore, one future challenge will be to find out how the RB hub of cell cycle control works in males, in order to fully understand the gender-specific evolution of the RB pathway in Volvox.
Abbreviations
| DP1 | = | dimerization partner 1 |
| CDK | = | cyclin-dependent kinase |
| RB | = | retinoblastoma |
| RBR | = | RB-related protein |
Figures and Tables
Figure 1 Simplified model for the activation of E2F1/DP1-specific target genes by phosphorylation of RBR1 in Volvox. (A) At inactive E2F1/DP1-specific target genes, the RB protein is found in a complex with the E2F1 transcription factor and its dimerization partner 1 (DP1), preventing E2F1/DP1 from activating transcription. (B) D-type cyclin is the regulatory subunit of a cyclin-dependent kinase/D-type cyclin holoenzyme. The D-type cyclin molecule has an LxCxE motif and binds to the LxCxE binding site of RBR1. (C) Hyperphosphorylation of RBR1 by the cyclin-dependent kinase/D-type cyclin holoenzyme results in the dissociation of RBR1 from the complex. The released E2F1/DP1 heterodimer promotes transcription of E2F1/DP1-specific target genes.
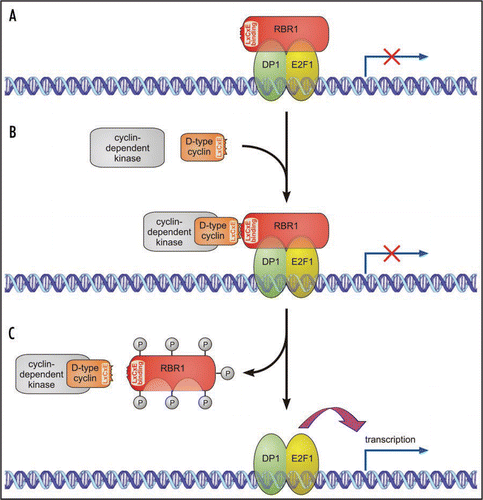
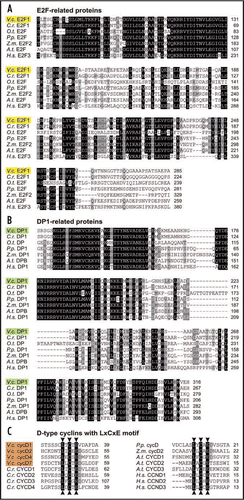
Figure 2 Protein sequence alignment of DP1, E2F1 and D-type cyclins of Volvox carteri with related proteins from green algae, moss, and higher plants and animals. Conserved amino acid residues were shaded using GeneDoc 2.6,Citation19 with similarity groups enabled. White letters on a black background represent residues conserved in >85% of the sequences at the corresponding position; residues shown in white letters on a gray background are conserved in >70% of the sequences; residues shown in black letters on a light gray background are conserved in >50% of the sequences. (A) E2F-related proteins: Volvox carteri E2F1 (V.c. E2F1, Acc. No. FJ898368), Chlamydomonas reinhardtii E2F1 (C.r. E2F1, Acc. No. EDP09526), Ostreococcus tauri E2F (O.t. E2F, Acc. No. AAV68605), Physcomitrella patens E2F (P.p. E2F, Acc. No. XP_001767806), Zea mays E2F2 (Z.m. E2F2, Acc. No. ACG37656), Arabidopsis thaliana E2F (A.t. E2F, Acc. No. BAB17029), and Homo sapiens E2F3 (H.s. E2F3, Acc. No. NP_001940). (B) DP1-related proteins: Volvox carteri DP1 (V.c. DP1, Acc. No. FJ898367), Chlamydomonas reinhardtii DP1 (C.r. DP1, Acc. No. XM_001700815), Ostreococcus tauri Dp (O.t. DP, Acc. No. AY675105), Physcomitrella patens DP1 (P.p. DP1, Acc. No. EDQ69967), Zea mays DP1 (Z.m. DP1, Acc. No. ACG47393), Arabidopsis thaliana DPB (A.t. DPB, Acc. No. NP_850757) and Homo sapiens Dp1 (H.s. Dp1, Acc. No. NP_009042). (C) LxCxE motifs of some D-type cyclins: Volvox carteri cycD1 (V.c. cycD1, JGI Volvox protein ID 127281), V. carteri cycD2 (V.c. cycD2, JGI Volvox protein ID 127277), V. carteri cycD4 (V.c. cycD4, JGI Volvox protein ID 127321), V. carteri cycD7 (V.c. cycD7, JGI Volvox protein ID 127283), Chlamydomonas reinhardtii CYCD1 (C.r. CYCD1, Acc. No. EDO98587), C. reinhardtii CYCD2 (C.r. CYCD2, Acc. No. EDP01778), C. reinhardtii CYCD3 (C.r. CYCD3, Acc. No. XM_001695417), C. reinhardtii CYCD4 (C.r. CYCD4, Acc. No. EDP00839), Physcomitrella patens cycD (P.p. cycD, Acc. No. CAD32542), Zea mays cycD2 (Z.m. cycD2, Acc. No. AAL83926), Arabidopsis thaliana CYCD1 (A.t. CYCD1, Acc. No. NM_105689), A. thaliana CYCD2 (A.t. CYCD2, Acc. No. NM_127815), A. thaliana CYCD3 (A.t. CYCD3, Acc. No. NP_195142), Homo sapiens CCND1 (H.s. CCND1, Acc. No. NM_053056), H. sapiens CCND2 (H.s. CCND2, Acc. No. NM_001759), and H. sapiens CCND3 (H.s. CCND3, Acc. No. NM_001760).
Addendum to:
References
- Weinberg RA. The retinoblastoma protein and cell cycle control. Cell 1995; 81:323 - 330
- Lundberg AS, Weinberg RA. Control of the cell cycle and apoptosis. Eur J Cancer 1999; 35:531 - 539
- Harbour JW, Dean DC. Rb function in cell cycle regulation and apoptosis. Nat Cell Biol 2000; 2:65 - 67
- Stevaux O, Dyson NJ. A revised picture of the E2F transcriptional network and RB function. Curr Opin Cell Biol 2002; 14:684 - 691
- Ben-Porath I, Weinberg RA. The signals and pathways activating cellular senescence. Int J Biochem Cell Biol 2005; 37:961 - 976
- Goodrich DW, Wang NP, Qian YW, Lee EY, Lee WH. The retinoblastoma gene product regulates progression through the G1 phase of the cell cycle. Cell 1991; 67:293 - 302
- Bartek J, Bartkova J, Lukas J. The retinoblastoma protein pathway and the restriction point. Curr Opin Cell Biol 1996; 8:805 - 814
- Du W, Pogoriler J. Retinoblastoma family genes. Oncogene 2006; 25:5190 - 5200
- Hanahan D, Weinberg RA. The hallmarks of cancer. Cell 2000; 100:57 - 70
- Dick FA. Structure-function analysis of the retinoblastoma tumor suppressor protein—is the whole a sum of its parts?. Cell Div 2007; 2:26
- Wikenheiser-Brokamp KA. Retinoblastoma family proteins: insights gained through genetic manipulation of mice. Cell Mol Life Sci 2006; 63:767 - 780
- Kianianmomeni A, Nematollahi G, Hallmann A. A gender-specific retinoblastoma-related protein in Volvox carteri implies a role for the retinoblastoma protein family in sexual development. Plant Cell 2008; 20:2399 - 2419
- Kirk DL. Volvox: Molecular-genetic Origins of Multicellularity and Cellular Differentiation. Developmental and Cell Biology Series 1998; Cambridge Cambridge University Press
- Kirk DL. Volvox as a model system for studying the ontogeny and phylogeny of multicellularity and cellular differentiation. J Plant Growth Regul 2000; 19:265 - 274
- Chellappan SP, Hiebert S, Mudryj M, Horowitz JM, Nevins JR. The E2F transcription factor is a cellular target for the RB protein. Cell 1991; 65:1053 - 1061
- Dyson N. The regulation of E2F by pRB-family proteins. Genes Dev 1998; 12:2245 - 2262
- Lee JO, Russo AA, Pavletich NP. Structure of the retinoblastoma tumour-suppressor pocket domain bound to a peptide from HPV E7. Nature 1998; 391:859 - 865
- Dahiya A, Gavin MR, Luo RX, Dean DC. Role of the LXCXE binding site in Rb function. Mol Cell Biol 2000; 20:6799 - 6805
- Nicholas KB, Nicholas HB, Deerfield DW. GeneDoc: Analysis and Visualization of Genetic Variation. Embnet News 1997; 4:14