Abstract
mDia proteins are members of the formin family of actin nucleating proteins that polymerize linear actin filaments. Such filaments form the core of thin, tubular, membrane-bound cell surface protrusions known as filopodia, which are a major feature of mammalian cell morphology. Filopodia are dynamic structures that help cells sense environmental cues, and play a role in cell migration, axon guidance, angiogenesis and other processes. RhoGTPases bind to and control the activity of mDia proteins, and several other binding partners of the three mDia1 isoforms—mDia1, mDia2 and mDia3—have been documented. Two independent pathways controlling mammalian filopodium formation have emerged, with one driven by the RhoGTPase Cdc42, and the other by Rif. While mDia2 has been the main formin implicated in forming filopodia, mDia1 has recently surfaced as the key formin utilized by both the Cdc42 and Rif pathways to drive filopodial protrusion.
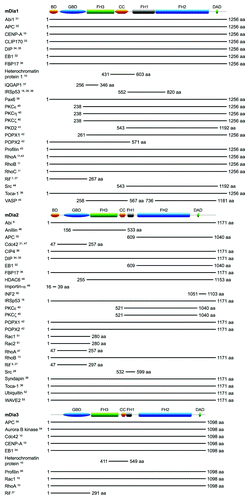
mDia Domain Organization and Interacting Proteins
When cells migrate, they extend dynamic membrane-bound actin-rich tubular protrusions known as filopodia.Citation1 Formins are a family of large multi-domain proteins that nucleate and polymerize actin to form linear actin filaments like those found within filopodia.Citation2 In this review we will focus on the formins mDia1–3 and their role in filopodium formation. Formins function as dimers and nucleate actin by means of a formin homology 2 (FH2) domain that binds globular actin monomers. Interaction of the adjacent formin homology 1 (FH1) domain with profilin effectively recruits actin monomers to the formin dimer, facilitating the polymerization process.Citation3 A subset of formins, known as Diaphanous-related formins (Drfs), bind to and are regulated by RhoGTPases.Citation3 Drfs are rendered inactive by interaction of a C-terminal Diaphanous autoregulatory domain (DAD) with an N-terminal Diaphanous inhibitory domain (DID) (). The binding of a RhoGTPase to the N-terminal GTPase binding domain (GBD) contributes to disruption of this autoinhibitory interaction, which results in the activation of the Drf.Citation2 In the case of the Drf FHOD1, phosphorylation of C-terminal serine and threonine residues by ROCK also overcomes the autoinhibition.Citation4 Other domains found in Drfs include a a dimerization domain (DD) and a coiled coil (CC) region, and some groups believe that the DD and DID together constitute a loosely defined formin homology 3 (FH3) domain.Citation3 An N-terminal phospholipid-binding basic domain (BD) has also been identified in the Drfs mDia1 and mDia2. mDia1 has three stretches of basic amino acids in this domain, which allows it to localize to the plasma membrane, while mDia2 has two.Citation5 In contrast, the Drf DAAM1 has only one stretch of basic amino acids in its N-terminal, and the distribution of constitutively active DAAM1 is restricted to the cytoplasm.Citation6 This might explain why only mDia1 and mDia2 have been implicated in the formation of membrane-bound cell surface protrusions like filopodia and lamellipodia,Citation7-Citation9 while mDia3—lacking this BD—has only been reported to generate cytoplasmic actin structures.Citation10 Several RhoGTPases and other proteins have been reported to bind to mDia1-3. These are summarized in .
Figure 1. Domain organization and interacting partners of mDia1-3. The shortest fragment(s) known to bind the respective mDia isoforms is shown for each interacting protein. Excluded from this diagram are YWK-II, which binds a 223 aa fragment of hDia1 that shares 96% aa sequence identity with mDia1 (903-1125 aa),Citation56 and INF2, which binds to aa 1181-1262 and aa 1051-1193 of what might be longer splice variants of mDia1 and mDia3 respectively.Citation50 Domain architecture diagrams were created using MyDomains Image Creator (prosite.expasy.org/cgi-bin/prosite/mydomains/).
mDia Proteins in Actin-Based Cellular Structures
mDia1-3 form several types of actin-based cellular structures. Within the cytoplasm, mDia1 gives rise to stress fibers,Citation11,Citation12 and mDia2 drives the actin dynamics that power vesicle movementCitation13 and creates the actin scaffold for constriction of the contractile ring during cytokinesis.Citation14 At the plasma membrane, both mDia1 and mDia2 have been shown to form lamellipodiaCitation8,Citation15 and filopodia.Citation7,Citation16 A role for formins in lamellipodial protrusion is not surprising—once thought to be structures comprised of dendritic networks of branched microfilaments, lamellipodia have recently been reported to contain linear actin filaments as well.Citation17 We have found mDia3 to be capable of inducing filopodia in N1E115 neuroblastoma cells, despite not being able to detect the presence of endogenous mDia3 protein in this particular cell line.Citation16 All three mDia isoforms have also been linked to invadopodia protrusion,Citation18 while mDia2 alone plays a role in forming the filopodial precursors of dendritic spines.Citation19 Actin dynamics leading to the formation of the phagocytic cup in macrophages are believed to involve mDia1 and mDia2 too.Citation20
mDia Proteins and Filopodium Formation in Mammalian Cells
Previous studies have pointed to a role for mDia2 in mammalian filopodia. A decrease in filopodial protrusion was seen in mouse fibroblasts overexpressing constitutively active Cdc42 and microinjected with anti-mDia2 antibodies, as well as cells cotransfected with activated Cdc42 and a non-functional mDia2 mutant.Citation21 In NIH3T3 fibroblasts, mDia2 localized to the tips of filopodia in cells overexpressing constitutively active Rif.Citation9 Constitutively active mDia2, when overexpressed alone in B16F1 melanoma cells, also accumulated at filopodial tips.Citation22 Furthermore, knockdown of mDia2 protein in mouse hippocampal neurons reduced the formation of the filopodial precursors of dendritic spines.Citation19 In more recent work, mDia1-3 have been shown to induce filopodia in neuronal cells when overexpressed on their own.Citation7,Citation16 However, only mDia1 was seen within filopodia, as observed by time lapse imaging of live cells.Citation7,Citation16 The lack of mDia2 and mDia3 in neuronal filopodia implies that these Drfs might be involved in the initiation of filopodium formation but not the elongation of the structures. One possibility is that mDia2 and mDia3 generate short microfilaments that are subsequently elongated by mDia1 to form mature filopodia. This would tie in with the suggestion that mDia2 is a relatively strong nucleator but poor elongator of microfilaments,Citation23 based on observations that it elongates microfilaments at a slower rate than mDia1.Citation24 The filopodia induced by full-length wildtype mDia2 appeared cylindrical and of even thickness along their length,Citation7,Citation16 and so did the filopodia formed by FH1FH2-mDia2, a fragment of mDia2 that consists of only the FH1 and FH2 domains.Citation8 This is unlike the club-shaped filopodia obtained by transfecting B16F1 cells with constitutively active mDia2,Citation8,Citation22 where the structures are packed with shorter microfilaments at their distal ends but contain relatively few long microfilaments that extend toward the base of the protrusions.Citation22 The high overexpression of constitutively active mDia2 could have resulted in endogenous mDia1 protein becoming a limiting factor on the process of filopodial microfilament elongation, giving rise to the club-shaped morphology of the filopodia. In addition, both full-length mDia2 and FH1FH2-mDia2 localized mostly to the cytoplasmCitation7,Citation8—in these experiments it is likely that less of the protein was present in filopodia, and there was enough endogenous mDia1 to elongate the smaller number of short microfilaments generated, thus resulting in filopodia of even thickness along their shafts. These interpretations of the findings would further implicate mDia1 as the key mDia isoform that elongates microfilaments to form mature filopodia. It would be interesting to see if knocking out mDia1 affects mDia2-driven filopodial protrusion—will the resulting mDia2-induced filopodia be shorter in length?
mDia1 was seen throughout the shafts of filopodia when overexpressed alone or together with IRSp53 or constitutively active Rif in neuronal cells.Citation7,Citation16 This is in contrast to the ‘tip nucleation’ model of filopodium formation,Citation1 where formins are expected to be found only at the tips of the protrusions. One possible explanation is that mDia1 dimers play a dual role in filopodium formation and are involved in not just polymerising the actin filaments but bundling them together as well. Another possibility is that filopodia consist of short, discontinuous actin filaments that do not span the entire length of the filopodial shaft. This has been shown by cryo-electron tomography studies to be the nature of the actin filaments that constitute Dictyostelium filopodia.Citation25 Superresolution microscopy studies would be able to reveal detail at the nanometre scale and help elucidate the specific locations of mDia1 and mDia2, as well as other proteins associated with filopodial protrusion, within the structures with much greater accuracy. This would help in establishing a better understanding of the roles of these proteins in the various stages of filopodium formation.
mDia Proteins in Filopodia Induced by Cdc42 and Rif
The Rho GTPases Cdc42 and Rif regulate distinct pathways to filopodium formation. Cdc42 works through IRSp53, which recruits to the plasma membrane the following proteins that modulate actin dynamics: N-WASP, Mena, WAVE2 and Eps8.Citation26 The Rif pathway to filopodia does not require IRSp53, N-WASP, Mena or WAVE2.Citation7 As for the mDia proteins, mDia1 appears to be the only isoform common to both pathways. In the filopodia of neuronal cells overexpressing IRSp53, mDia1 but not mDia2 was present, and was observed to interact with IRSp53 within the structures.Citation16 While both mDia1 and mDia2 were present in Rif filopodia, only mDia1 interacted with the RhoGTPase.Citation7 In addition, knockdown of either of these two isoforms resulted in a decrease in Rif-driven filopodium formation,Citation7 while IRSp53 filopodia were affected only by the silencing of mDia1 expression.Citation16 Taken together, it appears that mDia2 is not required for IRSp53 to form filopodia, and we have found that coexpressing mDia2 with IRSp53 leads to a loss of filopodia instead.Citation16 It remains to be seen as to why cells need two or even moreyetundiscovered pathways to form filopodia, and why IRSp53 requires only mDia1 when Rif appears to require both mDia1 and mDia2. Also, Rif has been shown to bind the GBD of mDia3, however the significance of this interaction has yet to be investigated.Citation27
mDia1Citation11,Citation27 and possibly mDia2Citation28 are involved in stress fiber formation in addition to filopodial protrusion. These two types of actin-based cellular structures appear to be linked—in fish fibroblasts, microfilaments in filopodia can become incorporated into stress fibers,Citation29 and it has been suggested that the reverse might occur in rat embryonic fibroblasts, with the actin freed up by the dissolution of stress fibers facilitating the protrusion of filopodia.Citation30 Rif interacts with both mDia1 and mDia2Citation27 and is able to trigger the formation of both filopodiaCitation7,Citation9 and stress fibers.Citation27 The dual role of these three proteins might point to a major role for them in controlling the balance between these two types of actin structures in cell migration.
Conclusions
It is clear that mDia proteins play an important role in mammalian filopodium formation. mDia2 appears to be specific for the Rif-mediated pathway whereas mDia1 is required for the pathways controlled by Cdc42 and Rif. It remains to be seen how exactly Rif utilizes two different formins, mDia1 and mDia2, to form filopodia. How Rif couples membrane deformation with actin dynamics to give rise to these structures has also yet to be resolved. Apart from mDia1 and mDia2, are there other proteins specific to the Rif pathway to filopodium formation? What potential roles do IRSp53 family proteins (IRTKS, MIM, ABBA and PinkBar) play in filopodial protrusion? These are some of the important questions to address in future studies on mammalian filopodia.
| Abbreviations: | ||
| BD | = | basic domain |
| CC | = | coiled coil |
| DAD | = | Diaphanous autoregulatory domain |
| DD | = | dimerization domain |
| DID | = | Diaphanous inhibitory domain |
| Drf | = | Diaphanous-related formin |
| FH1 | = | formin homology 1 |
| FH2 | = | formin homology 2 |
| FH3 | = | formin homology 3 |
| GBD | = | GTPase binding domain |
Acknowledgments
We would like to thank past and present members of the lab for their contribution to the research and to A-STAR for financial support.
References
- Mellor H. The role of formins in filopodia formation. Biochim Biophys Acta 2009; 1803:191-200.
- Young KG, Copeland JW. Formins in cell signaling. Biochim Biophys Acta 2010; 1803:183-90.
- Higgs HN. Formin proteins: a domain-based approach. Trends Biochem Sci 2005; 30:342 - 53; http://dx.doi.org/10.1016/j.tibs.2005.04.014; PMID: 15950879
- Takeya R, Taniguchi K, Narumiya S, Sumimoto H. The mammalian formin FHOD1 is activated through phosphorylation by ROCK and mediates thrombin-induced stress fibre formation in endothelial cells. EMBO J 2008; 27:618 - 28; http://dx.doi.org/10.1038/emboj.2008.7; PMID: 18239683
- Ramalingam N, Zhao H, Breitsprecher D, Lappalainen P, Faix J, Schleicher M. Phospholipids regulate localization and activity of mDia1 formin. Eur J Cell Biol 2010; 89:723 - 32; http://dx.doi.org/10.1016/j.ejcb.2010.06.001; PMID: 20619927
- Liu W, Sato A, Khadka D, Bharti R, Diaz H, Runnels LW, et al. Mechanism of activation of the Formin protein Daam1. Proc Natl Acad Sci U S A 2008; 105:210 - 5; http://dx.doi.org/10.1073/pnas.0707277105; PMID: 18162551
- Goh WI, Sudhaharan T, Lim KB, Sem KP, Lau CL, Ahmed S. Rif-mDia1 interaction is involved in filopodium formation independent of Cdc42 and Rac effectors. J Biol Chem 2011; 286:13681 - 94; http://dx.doi.org/10.1074/jbc.M110.182683; PMID: 21339294
- Yang C, Czech L, Gerboth S, Kojima S, Scita G, Svitkina T. Novel roles of formin mDia2 in lamellipodia and filopodia formation in motile cells. PLoS Biol 2007; 5:e317; http://dx.doi.org/10.1371/journal.pbio.0050317; PMID: 18044991
- Pellegrin S, Mellor H. The Rho family GTPase Rif induces filopodia through mDia2. Curr Biol 2005; 15:129 - 33; http://dx.doi.org/10.1016/j.cub.2005.01.011; PMID: 15668168
- Yasuda S, Oceguera-Yanez F, Kato T, Okamoto M, Yonemura S, Terada Y, et al. Cdc42 and mDia3 regulate microtubule attachment to kinetochores. Nature 2004; 428:767 - 71; http://dx.doi.org/10.1038/nature02452; PMID: 15085137
- Watanabe N, Kato T, Fujita A, Ishizaki T, Narumiya S. Cooperation between mDia1 and ROCK in Rho-induced actin reorganization. Nat Cell Biol 1999; 1:136 - 43; http://dx.doi.org/10.1038/11056; PMID: 10559899
- Hotulainen P, Lappalainen P. Stress fibers are generated by two distinct actin assembly mechanisms in motile cells. J Cell Biol 2006; 173:383 - 94; http://dx.doi.org/10.1083/jcb.200511093; PMID: 16651381
- Wallar BJ, Deward AD, Resau JH, Alberts AS. RhoB and the mammalian Diaphanous-related formin mDia2 in endosome trafficking. Exp Cell Res 2007; 313:560 - 71; http://dx.doi.org/10.1016/j.yexcr.2006.10.033; PMID: 17198702
- Watanabe S, Ando Y, Yasuda S, Hosoya H, Watanabe N, Ishizaki T, et al. mDia2 induces the actin scaffold for the contractile ring and stabilizes its position during cytokinesis in NIH 3T3 cells. Mol Biol Cell 2008; 19:2328 - 38; http://dx.doi.org/10.1091/mbc.E07-10-1086; PMID: 18287523
- Zaoui K, Honoré S, Isnardon D, Braguer D, Badache A. Memo-RhoA-mDia1 signaling controls microtubules, the actin network, and adhesion site formation in migrating cells. J Cell Biol 2008; 183:401 - 8; http://dx.doi.org/10.1083/jcb.200805107; PMID: 18955552
- Goh WI, Lim KB, Sudhaharan T, Sem KP, Bu W, Chou AM, et al. mDia1 and WAVE2 proteins interact directly with IRSp53 in filopodia and are involved in filopodium formation. J Biol Chem 2012; 287:4702 - 14; http://dx.doi.org/10.1074/jbc.M111.305102; PMID: 22179776
- Urban E, Jacob S, Nemethova M, Resch GP, Small JV. Electron tomography reveals unbranched networks of actin filaments in lamellipodia. Nat Cell Biol 2010; 12:429 - 35; http://dx.doi.org/10.1038/ncb2044; PMID: 20418872
- Lizárraga F, Poincloux R, Romao M, Montagnac G, Le Dez G, Bonne I, et al. Diaphanous-related formins are required for invadopodia formation and invasion of breast tumor cells. Cancer Res 2009; 69:2792 - 800; http://dx.doi.org/10.1158/0008-5472.CAN-08-3709; PMID: 19276357
- Hotulainen P, Llano O, Smirnov S, Tanhuanpää K, Faix J, Rivera C, et al. Defining mechanisms of actin polymerization and depolymerization during dendritic spine morphogenesis. J Cell Biol 2009; 185:323 - 39; http://dx.doi.org/10.1083/jcb.200809046; PMID: 19380880
- Colucci-Guyon E, Niedergang F, Wallar BJ, Peng J, Alberts AS, Chavrier P. A role for mammalian diaphanous-related formins in complement receptor (CR3)-mediated phagocytosis in macrophages. Curr Biol 2005; 15:2007 - 12; http://dx.doi.org/10.1016/j.cub.2005.09.051; PMID: 16303559
- Peng J, Wallar BJ, Flanders A, Swiatek PJ, Alberts AS. Disruption of the Diaphanous-related formin Drf1 gene encoding mDia1 reveals a role for Drf3 as an effector for Cdc42. Curr Biol 2003; 13:534 - 45; http://dx.doi.org/10.1016/S0960-9822(03)00170-2; PMID: 12676083
- Block J, Stradal TE, Hänisch J, Geffers R, Köstler SA, Urban E, et al. Filopodia formation induced by active mDia2/Drf3. J Microsc 2008; 231:506 - 17; http://dx.doi.org/10.1111/j.1365-2818.2008.02063.x; PMID: 18755006
- Faix J, Breitsprecher D, Stradal TE, Rottner K. Filopodia: Complex models for simple rods. Int J Biochem Cell Biol 2009; 41:1656 - 64; http://dx.doi.org/10.1016/j.biocel.2009.02.012; PMID: 19433307
- Kovar DR, Harris ES, Mahaffy R, Higgs HN, Pollard TD. Control of the assembly of ATP- and ADP-actin by formins and profilin. Cell 2006; 124:423 - 35; http://dx.doi.org/10.1016/j.cell.2005.11.038; PMID: 16439214
- Medalia O, Beck M, Ecke M, Weber I, Neujahr R, Baumeister W, et al. Organization of actin networks in intact filopodia. Curr Biol 2007; 17:79 - 84; http://dx.doi.org/10.1016/j.cub.2006.11.022; PMID: 17208190
- Lim KB, Bu W, Goh WI, Koh E, Ong SH, Pawson T, et al. The Cdc42 effector IRSp53 generates filopodia by coupling membrane protrusion with actin dynamics. J Biol Chem 2008; 283:20454 - 72; http://dx.doi.org/10.1074/jbc.M710185200; PMID: 18448434
- Fan L, Pellegrin S, Scott A, Mellor H. The small GTPase Rif is an alternative trigger for the formation of actin stress fibers in epithelial cells. J Cell Sci 2010; 123:1247 - 52; http://dx.doi.org/10.1242/jcs.061754; PMID: 20233848
- Tominaga T, Sahai E, Chardin P, McCormick F, Courtneidge SA, Alberts AS. Diaphanous-related formins bridge Rho GTPase and Src tyrosine kinase signaling. Mol Cell 2000; 5:13 - 25; http://dx.doi.org/10.1016/S1097-2765(00)80399-8; PMID: 10678165
- Nemethova M, Auinger S, Small JV. Building the actin cytoskeleton: filopodia contribute to the construction of contractile bundles in the lamella. J Cell Biol 2008; 180:1233 - 44; http://dx.doi.org/10.1083/jcb.200709134; PMID: 18362182
- Vetterkind S, Miki H, Takenawa T, Klawitz I, Scheidtmann KH, Preuss U. The rat homologue of Wiskott-Aldrich syndrome protein (WASP)-interacting protein (WIP) associates with actin filaments, recruits N-WASP from the nucleus, and mediates mobilization of actin from stress fibers in favor of filopodia formation. J Biol Chem 2002; 277:87 - 95; http://dx.doi.org/10.1074/jbc.M104555200; PMID: 11687573
- Ryu JR, Echarri A, Li R, Pendergast AM. Regulation of cell-cell adhesion by Abi/Diaphanous complexes. Mol Cell Biol 2009; 29:1735 - 48; http://dx.doi.org/10.1128/MCB.01483-08; PMID: 19158278
- Wen Y, Eng CH, Schmoranzer J, Cabrera-Poch N, Morris EJ, Chen M, et al. EB1 and APC bind to mDia to stabilize microtubules downstream of Rho and promote cell migration. Nat Cell Biol 2004; 6:820 - 30; http://dx.doi.org/10.1038/ncb1160; PMID: 15311282
- Lewkowicz E, Herit F, Le Clainche C, Bourdoncle P, Perez F, Niedergang F. The microtubule-binding protein CLIP-170 coordinates mDia1 and actin reorganization during CR3-mediated phagocytosis. J Cell Biol 2008; 183:1287 - 98; http://dx.doi.org/10.1083/jcb.200807023; PMID: 19114595
- Satoh S, Tominaga T. mDia-interacting protein acts downstream of Rho-mDia and modifies Src activation and stress fiber formation. J Biol Chem 2001; 276:39290 - 4; http://dx.doi.org/10.1074/jbc.M107026200; PMID: 11509578
- Eisenmann KM, Harris ES, Kitchen SM, Holman HA, Higgs HN, Alberts AS. Dia-interacting protein modulates formin-mediated actin assembly at the cell cortex. Curr Biol 2007; 17:579 - 91; http://dx.doi.org/10.1016/j.cub.2007.03.024; PMID: 17398099
- Aspenström P, Richnau N, Johansson AS. The diaphanous-related formin DAAM1 collaborates with the Rho GTPases RhoA and Cdc42, CIP4 and Src in regulating cell morphogenesis and actin dynamics. Exp Cell Res 2006; 312:2180 - 94; http://dx.doi.org/10.1016/j.yexcr.2006.03.013; PMID: 16630611
- Brandt DT, Marion S, Griffiths G, Watanabe T, Kaibuchi K, Grosse R. Dia1 and IQGAP1 interact in cell migration and phagocytic cup formation. J Cell Biol 2007; 178:193 - 200; http://dx.doi.org/10.1083/jcb.200612071; PMID: 17620407
- Fujiwara T, Mammoto A, Kim Y, Takai Y. Rho small G-protein-dependent binding of mDia to an Src homology 3 domain-containing IRSp53/BAIAP2. Biochem Biophys Res Commun 2000; 271:626 - 9; http://dx.doi.org/10.1006/bbrc.2000.2671; PMID: 10814512
- Tominaga T, Meng W, Togashi K, Urano H, Alberts AS, Tominaga M. The Rho GTPase effector protein, mDia, inhibits the DNA binding ability of the transcription factor Pax6 and changes the pattern of neurite extension in cerebellar granule cells through its binding to Pax6. J Biol Chem 2002; 277:47686 - 91; http://dx.doi.org/10.1074/jbc.M207539200; PMID: 12324464
- Eng CH, Huckaba TM, Gundersen GG. The formin mDia regulates GSK3beta through novel PKCs to promote microtubule stabilization but not MTOC reorientation in migrating fibroblasts. Mol Biol Cell 2006; 17:5004 - 16; http://dx.doi.org/10.1091/mbc.E05-10-0914; PMID: 16987962
- Rundle DR, Gorbsky G, Tsiokas L. PKD2 interacts and co-localizes with mDia1 to mitotic spindles of dividing cells: role of mDia1 IN PKD2 localization to mitotic spindles. J Biol Chem 2004; 279:29728 - 39; http://dx.doi.org/10.1074/jbc.M400544200; PMID: 15123714
- Xie Y, Tan EJ, Wee S, Manser E, Lim L, Koh CG. Functional interactions between phosphatase POPX2 and mDia modulate RhoA pathways. J Cell Sci 2008; 121:514 - 21; http://dx.doi.org/10.1242/jcs.013557; PMID: 18230650
- Watanabe N, Madaule P, Reid T, Ishizaki T, Watanabe G, Kakizuka A, et al. p140mDia, a mammalian homolog of Drosophila diaphanous, is a target protein for Rho small GTPase and is a ligand for profilin. EMBO J 1997; 16:3044 - 56; http://dx.doi.org/10.1093/emboj/16.11.3044; PMID: 9214622
- Yamana N, Arakawa Y, Nishino T, Kurokawa K, Tanji M, Itoh RE, et al. The Rho-mDia1 pathway regulates cell polarity and focal adhesion turnover in migrating cells through mobilizing Apc and c-Src. Mol Cell Biol 2006; 26:6844 - 58; http://dx.doi.org/10.1128/MCB.00283-06; PMID: 16943426
- Grosse R, Copeland JW, Newsome TP, Way M, Treisman R. A role for VASP in RhoA-Diaphanous signalling to actin dynamics and SRF activity. EMBO J 2003; 22:3050 - 61; http://dx.doi.org/10.1093/emboj/cdg287; PMID: 12805219
- Watanabe S, Okawa K, Miki T, Sakamoto S, Morinaga T, Segawa K, et al. Rho and anillin-dependent control of mDia2 localization and function in cytokinesis. Mol Biol Cell 2010; 21:3193 - 204; http://dx.doi.org/10.1091/mbc.E10-04-0324; PMID: 20660154
- Alberts AS, Bouquin N, Johnston LH, Treisman R. Analysis of RhoA-binding proteins reveals an interaction domain conserved in heterotrimeric G protein beta subunits and the yeast response regulator protein Skn7. J Biol Chem 1998; 273:8616 - 22; http://dx.doi.org/10.1074/jbc.273.15.8616; PMID: 9535835
- Destaing O, Saltel F, Gilquin B, Chabadel A, Khochbin S, Ory S, et al. A novel Rho-mDia2-HDAC6 pathway controls podosome patterning through microtubule acetylation in osteoclasts. J Cell Sci 2005; 118:2901 - 11; http://dx.doi.org/10.1242/jcs.02425; PMID: 15976449
- Miki T, Okawa K, Sekimoto T, Yoneda Y, Watanabe S, Ishizaki T, et al. mDia2 shuttles between the nucleus and the cytoplasm through the importin-alpha/beta- and CRM1-mediated nuclear transport mechanism. J Biol Chem 2009; 284:5753 - 62; http://dx.doi.org/10.1074/jbc.M806191200; PMID: 19117945
- Sun H, Schlondorff JS, Brown EJ, Higgs HN, Pollak MR. Rho activation of mDia formins is modulated by an interaction with inverted formin 2 (INF2). Proc Natl Acad Sci U S A 2011; 108:2933 - 8; http://dx.doi.org/10.1073/pnas.1017010108; PMID: 21278336
- Ji P, Jayapal SR, Lodish HF. Enucleation of cultured mouse fetal erythroblasts requires Rac GTPases and mDia2. Nat Cell Biol 2008; 10:314 - 21; http://dx.doi.org/10.1038/ncb1693; PMID: 18264091
- DeWard AD, Alberts AS. Ubiquitin-mediated degradation of the formin mDia2 upon completion of cell division. J Biol Chem 2009; 284:20061 - 9; http://dx.doi.org/10.1074/jbc.M109.000885; PMID: 19457867
- Beli P, Mascheroni D, Xu D, Innocenti M. WAVE and Arp2/3 jointly inhibit filopodium formation by entering into a complex with mDia2. Nat Cell Biol 2008; 10:849 - 57; http://dx.doi.org/10.1038/ncb1745; PMID: 18516090
- Cheng L, Mao Y. mDia3-EB1-APC: A connection between kinetochores and microtubule plus ends. Commun Integr Biol 2011; 4:480 - 2; PMID: 21966577
- Behnen M, Murk K, Kursula P, Cappallo-Obermann H, Rothkegel M, Kierszenbaum AL, et al. Testis-expressed profilins 3 and 4 show distinct functional characteristics and localize in the acroplaxome-manchette complex in spermatids. BMC Cell Biol 2009; 10:34; http://dx.doi.org/10.1186/1471-2121-10-34; PMID: 19419568
- Zhang SM, Miao SY, Wang LF, Koide SS. Evidence for the binding of a human sperm component with diaphanous protein. Arch Androl 2001; 46:29 - 35; http://dx.doi.org/10.1080/01485010150211128; PMID: 11204614