Abstract
The regulated synthesis of specific proteins at the synapse is important for neuron plasticity, and several localized mRNAs are translated upon specific stimulus. Repression of mRNA translation is linked to the formation of mRNA-silencing foci, including Processing Bodies (PBs) and Stress Granules (SGs), which are macromolecular aggregates that harbor silenced messengers and associated proteins. In a recent work, we identified a kind of mRNA-silencing foci unique to neurons, termed S-foci, that contain the post-transcriptional regulator Smaug1/SAMD4. Upon specific synaptic stimulation, the S-foci dissolve and release mRNAs to allow their translation, paralleling the cycling of mRNAs between PBs and polysomes in other cellular contexts. Smaug 1 and other proteins involved in mRNA regulation in neurons contain aggregation domains distinct from their RNA binding motifs, and we speculate that self-aggregation helps silencing and transport. In addition to S-foci and PBs, other foci formed by distinct RNA binding proteins, such as TDP-43 and FMRP among others, respond dynamically to specific synaptic stimuli. We propose the collective name of synaptic activity-regulated mRNA silencing (SyAS) foci for these RNP aggregates that selectively respond to distinct stimulation patterns and contribute to the fine-tuning of local protein synthesis at the synapse.
Local translation at the synapse is key for neuronal activity. Hundreds of mRNAs encoding highly diverse proteins involved in distinct structures and processes are transported from the cell soma to the dendrites and synapses with distinct localization patterns. Adding significance to this exquisite spatial distribution, the translation of these transcripts is regulated by synaptic activity, thereby providing a precision mechanism for the local production of specific proteins in response to local changes. Dendritically localized mRNA molecules may remain silent most of the time, and become active upon specific stimulus.Citation1 An emerging concept is that mRNA repression is linked to the formation of mRNA-silencing foci. Processing Bodies (PBs) and Stress Granules (SGs) are the founding members of this novel family of cellular structures, which are cytoplasmic supramolecular aggregates of repressed RNA and inhibitory proteins. PBs are ubiquitous and SGs are specific of the stress response. SGs are rapidly assembled during the global translational silencing triggered by cell stress. Both PBs and SGs are dynamic and dissolve when the associated mRNAs engage in translation according to cellular needs.Citation2-Citation4
Recently, we identified a mRNA-silencing foci specific of neurons. These foci contain Smaug1/SAMD4, a translational repressor initially identified in the Drosophila embryo, and that we found expressed in the mammalian CNS.Citation5,Citation6 Smaug proteins are novel mRNA regulators and they recognize their targets by a unique strategy, involving a SAM domain that binds a number of related motifs collectively termed SRE (Smaug Recognition Element).Citation7 Smaug1 foci are distinct from PBs and other neuronal RNA granules hitherto described, and were named S-foci. Both in neurons as well as in transfected cell lines, the integrity of the S-foci inversely correlates with that of polysomes, as expected for mRNA-silencing foci. The S-foci dissolve when mRNAs are trapped into polysomes. Conversely, S-foci formation is enhanced when mRNAs are released from polysomes. Drosophila Smaug also forms mRNA silencing foci when expressed in mammalian cells.Citation5,Citation6
The S-foci are present at dendrites and remarkably associated to the post-synapses. Up to 60% of the dendritic spines of mature hippocampal neurons contain S-foci. An important finding is that these neuron-specific mRNA silencing foci respond to synaptic activation. The S-foci dissolve and release target mRNAs to allow their translation upon stimulation of the N-Methyl D-aspartate (NMDA) receptor. The response is rapid and maximal dissolution occurs between 1 to 5 min after stimulation. The effect is transient and after massive dissolution the S-foci begin to reassemble, provided that polysomes are allowed to disengage after the translation pulse. These observations suggest that mRNAs cycle between S-foci and polysomes, thus paralleling the cycling of mRNAs between PBs and polysomes in other cellular contexts ().Citation6
Figure 1. SyAS-foci and translational regulation at the synapse. (A) Several mRNA-silencing foci, including S-foci, PBs and FMRP granules, among others, are present in dendrites and dendritic spines. S-foci are different from FMRP granules and PBs. The three kind of SyAS-foci may coexist in a single dendritic spine. (B-D) Distinct SyAS-foci respond to specific stimuli, dissolving and releasing specific mRNAs. Stimulation of NMDAR or mGlutR affect S-foci and FMRP granules, and activates the translation of CamKIIα, among others.6 About half of synapse-localized CamKIIα mRNA is associated to S-foci under resting conditions and is released upon NMDA stimulation and S-foci dissolution.6 In addition, CamKIIα mRNA is as well regulated by FMRP and PBs,30 opening the possibility of multiple regulation by distinct pathways. NMDAR stimulation provokes a global silencing, and the translation of a number of transcripts, including Kv1.1 mRNA among others, is repressed.6,36
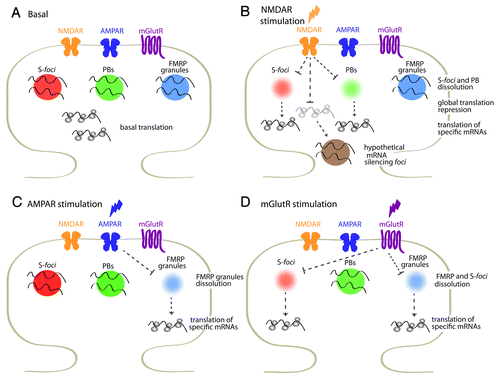
The S-foci apparently do not respond to α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid (AMPA) receptor stimulation. The S-foci enter a round of relaxation and re-assembly upon stimulation of metabotropic glutamate receptors (mGlutR), with a distinctive time-course, thus allowing the transient release of mRNAs followed by translation (Luchelli and Boccaccio, unpublished) ().Citation6 Both NMDAR and mGlutR activation trigger the local synthesis of a number of proteins involved in synapse remodelling, including CamKIIα, a key signaling molecule.Citation8 Current evidence indicates that the mRNA encoding CamKIIα is associated to S-foci and released upon NMDAR activation. Molecular abrogation of Smaug1 by RNAi strategies leads to a dramatic alteration of dendritic spine morphology and excitability. Deregulation of CamKIIα mRNA undoubtedly contributes to these defects.Citation6 In addition, we speculate that Smaug1 regulates other mRNAs. Supporting this notion, the Drosophila molecule controls the stability and translation of hundreds of maternal mRNAs during early development, and the SAM domain accommodates a number of variations on the cognate RNA motif.Citation7,Citation9-Citation13 A profound remodelling of the transcriptome occurs during neuron development and synapse formation. A large number of neuronal mRNAs encoding different cell functions and that are expressed early during neuron maturation show a decrease after the onset of synaptogenesis.Citation14 Thus, paralleling the role of Drosophila Smaug as a major determinant of the maternal-to-zygotic transition, we speculate that mammalian Smaug1 is a major mRNA regulator during synaptogenesis, a stage when Smaug1 expression is maximal.Citation6 Among other candidates, nanos 1—homologous to Drosophila nanos, which is the best characterized Smaug target in the fly—is likely to be regulated by mammalian Smaug1. Drosophila nanos is under Smaug control in the embryo and in peripheral neurons, where it affects the dendritic arbour.Citation15-Citation17 Nanos is also a translational repressor, and is recruited to mRNAs by its partner Pumilio. Both in mammalian and fly neurons, Pumilio affects dendritogenesis and/or synaptogenesis.Citation15,Citation16,Citation18-Citation21 Thus, the relevance of the Smaug -Nanos/Pumilio post-transcriptional regulatory pathway is likely to be conserved from the insect to the mammalian CNS.
The molecular mechanism for mRNA repression by mammalian Smaug1 is unknown, but is expected to be similar to that of Drosophila Smaug, which operates at several levels. Drosophila Smaug blocks translation initiation and induces mRNA decay by triggering deadenylation.Citation10-Citation12,Citation22 Both mechanisms are likely to occur in the mammalian counterpart. Deadenylation is frequent in neuronal mRNAs, and the mRNA encoding CamKIIα, which is repressed in S-foci, is activated by cytoplasmic polyadenylation.
Whether aggregation of Smaug1 in discrete structures is required for efficient mRNA repression is unknown. Several proteins involved in general mRNA silencing have distinct oligomerization domains.Citation2-Citation4 Among others motifs, regions enriched in glutamine (Q) and asparagine (N), which have a propensity to assemble into ordered aggregates, are frequent in PB components.Citation2-Citation4 None of these oligomerization domains are present in Smaug1. We found that Smaug1 aggregation is independent of RNA binding and of the presence of PBs.Citation6 All this indicates that S-foci formation is not a consequence of mRNA repression, and allows the speculation that aggregation is important for Smaug1 function. Supporting this, granule formation is conserved in Drosophila Smaug. Like Smaug1 in hippocampal neurons, the fly molecule is found in foci in the embryoCitation22 and in large RNP complexes that can be isolated biochemically.Citation23 In addition, Drosophila Smaug forms foci when expressed in mammalian cells.Citation5 Self-aggregation is a common theme in several RNA binding proteins relevant to RNA regulation in neurons. Drosophila FMRP has a Q/N rich domain that mediates protein-protein interaction and that is important for the establishment of short-term memory.Citation24 The active form of Cytoplasmic Polyadenilation Element Binding protein (CPEB) from Aplysia forms small aggregates that bind CPE-containing mRNAs, thus stimulating their cytoplasmic polyadenylation.Citation25 Orb2, the Drosophila homolog, forms amyloid-like oligomers and neurons with Orb2 oligomers are the site for long-term memory storage.Citation26 RNG105, a post-transcriptional repressor found in dendritic RNA granules bears a coiled-coil region that is necessary for both translational repression and aggregation. Loss of RNG105 results in aberrant synapse formation and degeneration of neuronal networks.Citation27 Mammalian Pumilio forms granules in both neurons and cell lines,Citation19 and Drosophila Pumilio and its nematode ortolog PUF-9 localize to microscopically visible aggregates when expressed in yeast cells.Citation21 Pumilio bears a Q/N rich segment at the N-terminus. This prion-like region is important for recruitment of mammalian Pumilio 2 to SGsCitation19 and regulates postsynaptic Pumilio function in the fly.Citation21 Messenger RNAs translated at the synapses travel long distances from the cell soma, and they move packaged in granules. We speculate that self-aggregation of Smaug1 and other translational repressors may facilitate the transport of their target mRNAs.
Simultaneously with the activation of mRNAs stored in the S-foci, we and others have shown that NMDAR stimulation triggers a global translational silencing.Citation1,Citation6 Using the FUNCAT strategy,Citation28 we found that in a few minutes, the protein synthesis rate in the surroundings of the stimulated synapses is reduced to half of the normal values (). This rapid translation shut down is akin to the acute translation repression upon cellular stress, and whether it involves the assembly of specific mRNA silencing foci remains to be investigated. SGs form in neurons immediately upon stress induction, but not upon NMDAR stimulationCitation6 (Luchelli and Boccaccio, unpublished), leaving the point unanswered.
In addition to the S-foci, which are exclusive of mature neurons, PBs also appears to play a role in translation regulation upon synaptic activity. Neuronal PBs diverge from those present in cell lines, and PB components that usually colocalize in a single kind of foci in cell lines are found in separate foci in dendrites.Citation29-Citation31 Synaptic activity affects the dynamics and integrity of dendritic PBs,Citation31-Citation33 and the speculation is made that this allows the release of mRNAs and their incorporation into polysomes. PBs and several neuronal RNA granules containing FMRP (FragileX Mental Retardation Protein), FUS/TLS (fused in sarcoma/translated in liposarcoma), TDP43 (TAR DNA-binding protein 43), Staufen, Pumilio, ZBP1 (zip code-binding protein 1) and others, are motile and believed to contribute to RNA transport. We anticipate that many of them will behave as mRNA-silencing foci and will respond to synaptic stimulation with enhanced assembly or dissolution to control the availability of specific transcripts. We propose the name of synaptic-activity regulated mRNA silencing foci (SyAS-foci) to these structures. Supporting these speculations, in addition to S-foci and PBs, the dynamics of a number of RNA granules is affected by neuronal activity. FMRP forms granules that dissolve upon AMPAR stimulation, which does not affect the integrity of the S-foci.Citation6,Citation34 Depolarization enhances TDP43 granules in dendrites.Citation35 RNG105 granules shrink and DCP1a-containing PBs are enhanced upon BDNF exposure.Citation32 All this suggests that local translation upon stimulation by specific neurotransmitters or neurotrofins involves the concerted regulation of distinct SyAS-foci ().
Our recent results show that Smaug1 knockdown severely disrupts spine morphology and neuron excitability, thus resembling the phenotype associated to Fragil X Mental Retardation Syndrome, a serious neurological disease provoked by altered expression of FMRP. Deregulation of Pumilio, Staufen or RNG105 provokes similar effects.Citation2,Citation18,Citation27 S-foci are distinct from the RNA granules containing these molecules, suggesting that Smaug1 and these non-related RNA binding proteins act in parallel pathways. Collectively, these observations emphasize the relevance of translation regulation by a plethora of SyAS-foci in neuron development and homeostasis.
| Abbreviations: | ||
| AMPA | = | α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid |
| CamKII | = | Ca2+/calmodulin-dependent protein kinases II |
| CPEB | = | cytoplasmic polyadenylation element binding protein |
| FMRP | = | fragile X mental retardation protein |
| FUS/TLS | = | fused in sarcoma/translated in liposarcoma |
| mGlutR | = | metabotropic glutamate receptor, NMDA, N-Methyl D-aspartic acid |
| PBs | = | processing bodies |
| RBP | = | RNA-binding protein |
| RNG105 | = | RNA granule protein 105 |
| RNP | = | ribonucleoprotein |
| S-foci | = | Smaug1 foci |
| SGs | = | stress granules |
| SyAS foci | = | synaptic activity-regulated mRNA silencing foci |
| TDP43 | = | TAR DNA-binding protein 43 |
| ZBP1 | = | zip code-binding protein 1 |
Acknowledgments
We are grateful to late David R. Colman for constant support and advice, and to Lorena Benseñor for critical reading of this manuscript. This work was supported by the following grants: UBACyT X311 from University of Buenos Aires, Argentina; PIP 6173 from Consejo Nacional de Investigaciones Científicas y Tecnológicas (CONICET); PICT 38006 and PICT 1965 from Agencia Nacional de Promoción Científica y Tecnológica, (ANPCyT), Argentina.
References
- Schuman EM, Dynes JL, Steward O. Synaptic regulation of translation of dendritic mRNAs. J Neurosci 2006; 26:7143 - 6; http://dx.doi.org/10.1523/JNEUROSCI.1796-06.2006; PMID: 16822969
- Thomas MG, Loschi M, Desbats MA, Boccaccio GL. RNA granules: the good, the bad and the ugly. Cell Signal 2011; 23:324 - 34; http://dx.doi.org/10.1016/j.cellsig.2010.08.011; PMID: 20813183
- Buchan JR, Parker R. Eukaryotic stress granules: the ins and outs of translation. Mol Cell 2009; 36:932 - 41; http://dx.doi.org/10.1016/j.molcel.2009.11.020; PMID: 20064460
- Franks TM, Lykke-Andersen J. The control of mRNA decapping and P-body formation. Mol Cell 2008; 32:605 - 15; http://dx.doi.org/10.1016/j.molcel.2008.11.001; PMID: 19061636
- Baez MV, Boccaccio GL. Mammalian Smaug is a translational repressor that forms cytoplasmic foci similar to stress granules. J Biol Chem 2005; 280:43131 - 40; http://dx.doi.org/10.1074/jbc.M508374200; PMID: 16221671
- Baez MV, Luchelli L, Maschi D, Habif M, Pascual M, Thomas MG, et al. Smaug1 mRNA-silencing foci respond to NMDA and modulate synapse formation. J Cell Biol 2011; 195:1141 - 57; http://dx.doi.org/10.1083/jcb.201108159; PMID: 22201125
- Aviv T, Lin Z, Ben-Ari G, Smibert CA, Sicheri F. Sequence-specific recognition of RNA hairpins by the SAM domain of Vts1p. Nat Struct Mol Biol 2006; 13:168 - 76; http://dx.doi.org/10.1038/nsmb1053; PMID: 16429151
- Lisman J, Yasuda R, Raghavachari S. Mechanisms of CaMKII action in long-term potentiation. Nat Rev Neurosci 2012; 13:169 - 82; PMID: 22334212
- Benoit B, He CH, Zhang F, Votruba SM, Tadros W, Westwood JT, et al. An essential role for the RNA-binding protein Smaug during the Drosophila maternal-to-zygotic transition. Development 2009; 136:923 - 32; http://dx.doi.org/10.1242/dev.031815; PMID: 19234062
- Nelson MR, Leidal AM, Smibert CA. Drosophila Cup is an eIF4E-binding protein that functions in Smaug-mediated translational repression. EMBO J 2004; 23:150 - 9; http://dx.doi.org/10.1038/sj.emboj.7600026; PMID: 14685270
- Semotok JL, Cooperstock RL, Pinder BD, Vari HK, Lipshitz HD, Smibert CA. Smaug recruits the CCR4/POP2/NOT deadenylase complex to trigger maternal transcript localization in the early Drosophila embryo. Curr Biol 2005; 15:284 - 94; http://dx.doi.org/10.1016/j.cub.2005.01.048; PMID: 15723788
- Semotok JL, Luo H, Cooperstock RL, Karaiskakis A, Vari HK, Smibert CA, et al. Drosophila maternal Hsp83 mRNA destabilization is directed by multiple SMAUG recognition elements in the open reading frame. Mol Cell Biol 2008; 28:6757 - 72; http://dx.doi.org/10.1128/MCB.00037-08; PMID: 18794360
- Tadros W, Goldman AL, Babak T, Menzies F, Vardy L, Orr-Weaver T, et al. SMAUG is a major regulator of maternal mRNA destabilization in Drosophila and its translation is activated by the PAN GU kinase. Dev Cell 2007; 12:143 - 55; http://dx.doi.org/10.1016/j.devcel.2006.10.005; PMID: 17199047
- Valor LM, Charlesworth P, Humphreys L, Anderson CN, Grant SG. Network activity-independent coordinated gene expression program for synapse assembly. Proc Natl Acad Sci U S A 2007; 104:4658 - 63; http://dx.doi.org/10.1073/pnas.0609071104; PMID: 17360580
- Menon KP, Sanyal S, Habara Y, Sanchez R, Wharton RP, Ramaswami M, et al. The translational repressor Pumilio regulates presynaptic morphology and controls postsynaptic accumulation of translation factor eIF-4E. Neuron 2004; 44:663 - 76; http://dx.doi.org/10.1016/j.neuron.2004.10.028; PMID: 15541314
- Ye B, Petritsch C, Clark IE, Gavis ER, Jan LY, Jan YN. Nanos and Pumilio are essential for dendrite morphogenesis in Drosophila peripheral neurons. Curr Biol 2004; 14:314 - 21; PMID: 14972682
- Brechbiel JL, Gavis ER. Spatial regulation of nanos is required for its function in dendrite morphogenesis. Curr Biol 2008; 18:745 - 50; http://dx.doi.org/10.1016/j.cub.2008.04.033; PMID: 18472422
- Vessey JP, Schoderboeck L, Gingl E, Luzi E, Riefler J, Di Leva F, et al. Mammalian Pumilio 2 regulates dendrite morphogenesis and synaptic function. Proc Natl Acad Sci U S A 2010; 107:3222 - 7; http://dx.doi.org/10.1073/pnas.0907128107; PMID: 20133610
- Vessey JP, Vaccani A, Xie Y, Dahm R, Karra D, Kiebler MA, et al. Dendritic localization of the translational repressor Pumilio 2 and its contribution to dendritic stress granules. J Neurosci 2006; 26:6496 - 508; http://dx.doi.org/10.1523/JNEUROSCI.0649-06.2006; PMID: 16775137
- Menon KP, Andrews S, Murthy M, Gavis ER, Zinn K. The translational repressors Nanos and Pumilio have divergent effects on presynaptic terminal growth and postsynaptic glutamate receptor subunit composition. J Neurosci 2009; 29:5558 - 72; http://dx.doi.org/10.1523/JNEUROSCI.0520-09.2009; PMID: 19403823
- Salazar AM, Silverman EJ, Menon KP, Zinn K. Regulation of synaptic Pumilio function by an aggregation-prone domain. J Neurosci 2010; 30:515 - 22; http://dx.doi.org/10.1523/JNEUROSCI.2523-09.2010; PMID: 20071514
- Rouget C, Papin C, Boureux A, Meunier AC, Franco B, Robine N, et al. Maternal mRNA deadenylation and decay by the piRNA pathway in the early Drosophila embryo. Nature 2010; 467:1128 - 32; http://dx.doi.org/10.1038/nature09465; PMID: 20953170
- Jeske M, Moritz B, Anders A, Wahle E. Smaug assembles an ATP-dependent stable complex repressing nanos mRNA translation at multiple levels. EMBO J 2011; 30:90 - 103; http://dx.doi.org/10.1038/emboj.2010.283; PMID: 21081899
- Banerjee P, Schoenfeld BP, Bell AJ, Choi CH, Bradley MP, Hinchey P, et al. Short- and long-term memory are modulated by multiple isoforms of the fragile X mental retardation protein. J Neurosci 2010; 30:6782 - 92; http://dx.doi.org/10.1523/JNEUROSCI.6369-09.2010; PMID: 20463240
- Si K, Lindquist S, Kandel ER. A neuronal isoform of the aplysia CPEB has prion-like properties. Cell 2003; 115:879 - 91; http://dx.doi.org/10.1016/S0092-8674(03)01020-1; PMID: 14697205
- Majumdar A, Cesario WC, White-Grindley E, Jiang H, Ren F, Khan MR, et al. Critical role of amyloid-like oligomers of Drosophila Orb2 in the persistence of memory. Cell 2012; 148:515 - 29; http://dx.doi.org/10.1016/j.cell.2012.01.004; PMID: 22284910
- Shiina N, Yamaguchi K, Tokunaga M. RNG105 deficiency impairs the dendritic localization of mRNAs for Na+/K+ ATPase subunit isoforms and leads to the degeneration of neuronal networks. J Neurosci 2010; 30:12816 - 30; http://dx.doi.org/10.1523/JNEUROSCI.6386-09.2010; PMID: 20861386
- Dieterich DC, Hodas JJ, Gouzer G, Shadrin IY, Ngo JT, Triller A, et al. In situ visualization and dynamics of newly synthesized proteins in rat hippocampal neurons. Nat Neurosci 2010; 13:897 - 905; http://dx.doi.org/10.1038/nn.2580; PMID: 20543841
- di Penta A, Mercaldo V, Florenzano F, Munck S, Ciotti MT, Zalfa F, et al. Dendritic LSm1/CBP80-mRNPs mark the early steps of transport commitment and translational control. J Cell Biol 2009; 184:423 - 35; http://dx.doi.org/10.1083/jcb.200807033; PMID: 19188494
- Hillebrand J, Pan K, Kokaram A, Barbee S, Parker R, Ramaswami M. The Me31B DEAD-box helicase localizes to postsynaptic foci and regulates expression of a CaMKII reporter mRNA in dendrites of Drosophila olfactory projection neurons. Front Neural Circuits 2010; 4:121; http://dx.doi.org/10.3389/fncir.2010.00121; PMID: 21267420
- Cougot N, Bhattacharyya SN, Tapia-Arancibia L, Bordonné R, Filipowicz W, Bertrand E, et al. Dendrites of mammalian neurons contain specialized P-body-like structures that respond to neuronal activation. J Neurosci 2008; 28:13793 - 804; http://dx.doi.org/10.1523/JNEUROSCI.4155-08.2008; PMID: 19091970
- Huang YW, Ruiz CR, Eyler EC, Lin K, Meffert MK. Dual regulation of miRNA biogenesis generates target specificity in neurotrophin-induced protein synthesis. Cell 2012; 148:933 - 46; http://dx.doi.org/10.1016/j.cell.2012.01.036; PMID: 22385959
- Zeitelhofer M, Karra D, Macchi P, Tolino M, Thomas S, Schwarz M, et al. Dynamic interaction between P-bodies and transport ribonucleoprotein particles in dendrites of mature hippocampal neurons. J Neurosci 2008; 28:7555 - 62; http://dx.doi.org/10.1523/JNEUROSCI.0104-08.2008; PMID: 18650333
- Antar LN, Afroz R, Dictenberg JB, Carroll RC, Bassell GJ. Metabotropic glutamate receptor activation regulates fragile x mental retardation protein and FMR1 mRNA localization differentially in dendrites and at synapses. J Neurosci 2004; 24:2648 - 55; http://dx.doi.org/10.1523/JNEUROSCI.0099-04.2004; PMID: 15028757
- Wang IF, Wu LS, Chang HY, Shen CK. TDP-43, the signature protein of FTLD-U, is a neuronal activity-responsive factor. J Neurochem 2008; 105:797 - 806; http://dx.doi.org/10.1111/j.1471-4159.2007.05190.x; PMID: 18088371
- Raab-Graham KF, Haddick PC, Jan YN, Jan LY. Activity- and mTOR-dependent suppression of Kv1.1 channel mRNA translation in dendrites. Science 2006; 314:144 - 8; http://dx.doi.org/10.1126/science.1131693; PMID: 17023663