Abstract
Eusocial Hymenoptera, such as the European honey bee, Apis mellifera, have the highest recombination rates of multicellular animals.Citation1 Recently, we showedCitation2 that a side-effect of recombination in the honey bee, GC biased gene conversion (bGC), helps maintain the unusual bimodal GC-content distribution of the bee genome by increasing GC-content in high recombination areas while low recombination areas are losing GC-content because of biased AT mutations and low rates of bGC. Although the very high recombination rate of A. mellifera makes GC-content evolution easier to study, the pattern is consistent with results found in many other species including mammals and yeast.Citation3 Also consistent across phyla is the association of higher genetic diversity and divergence with high GC and high recombination areas.Citation4,Citation5 Finally, we showed that genes overexpressed in the brains of workers cluster in GC-rich genomic areas with the highest rates of recombination and molecular evolution.Citation2 In this Addendum we present a conceptual model of how eusociality and high recombination rates may co-evolve.
Text
Recombination is high in several species with independent origins of eusociality.Citation1,Citation6,Citation7 A number of studies have demonstrated fitness benefits in eusocial insects due to increased genetic variation in the colony,Citation8-Citation10 and it was hypothesized that high recombination rates in eusocial insects were selected for because recombination increases worker genotypic diversity.Citation1,Citation11 However, a recent modeling study demonstrated that recombination has little short-term impact on within-colony variation compared with factors such as polyandry.Citation12 What conditions favor the evolution of high recombination rates in social insects? Theoretical and empirical work points to two major factors: small effective population size, and strong selection on linked genes.Citation13,Citation14
Consider the early stages of the evolution of eusociality. First, the evolution of sociality is expected to greatly reduce effective population sizeCitation15 because of the larger amount of resources required to raise both workers and reproductives, higher variance in reproductive output between colonies and worker-produced males.Citation16-Citation18 The reduction in effective population size associated with the evolution of eusociality is expected to both increase linkage disequilibriumCitation19 and increase the frequency of slightly deleterious mutations via drift.Citation20 When recombination rate (R) is low, new advantageous mutations are less likely to fix if nearby deleterious alleles hitchhike along with themCitation21—a phenomenon called Hill-Robertson interference.Citation22 Low R will also slow selection on advantageous alleles that are physically proximate but on different haplotypes, as the mutation with the highest selection coefficient will tend to drive the others to extinction before recombination can place them on the same haplotype.Citation23 The initial stages of the evolution of eusociality can thus be characterized by lower Ne, high linkage disequilibrium, higher genetic loads and reduced effectiveness of natural selection due to interference between linked mutations.
The above arguments suggest a paradoxical conclusion; the transition to eusociality created sub-optimal conditions for the evolution of caste-specialization and worker evolution via natural selection. However, the concurrent evolution of high recombination rates with eusociality—as observed across a number of independently derived eusocial lineagesCitation1—can reduce interference between linked mutations, thereby greatly enhancing the effectiveness of natural selection. Assuming heritable variation in recombination rates,Citation24,Citation25 queens with high rates of recombination are expected to produce more recombinant gametes allowing natural selection to act more independently on physically linked mutations. This is expected to increase both queen and worker fitness by reducing interference and enhancing natural selection, which, in turn, should further select for higher rates of recombination.
We propose three non-mutually exclusive processes that can exert strong selective pressures for high recombination rates in eusocial insects.
Divergence of Queen and Worker Phenotypes
Caste specialization and division of labor is one of the hallmarks of eusociality. The genomes of social insects must generate two (or more) female phenotypes from the same genome (i.e., queens and workers). Several non-mutually exclusive processes can facilitate phenotypic divergence of castes, including caste-specific gene expression, alternative splicing, methylation and gene duplication.Citation26-Citation28 However, the end-result of the above processes is often very similar: mutations affecting worker phenotypes are likely to be physically proximate to mutations affecting queen phenotypes. This will be true in cases where the same genes are expressed in both queens and workers but at different rates or at different times,Citation29-Citation31 or in cases with caste-specific splice formsCitation32 and or caste-specific methylation patterns,Citation32 or if gene duplication events lead to physically proximate caste-specific genes. In the above cases, we expect a general tug-of-war between linked mutations influencing worker and queen traits.Citation33,Citation34 How is this tug-of-war resolved? Recent theory suggests that, all other effects being equal, queens invariably win because selection acts on them directly, but workers only experience indirect selection.Citation35,36 High recombination rates are expected to reduce the tug-of-war between queens and workers, thereby allowing natural selection to optimize caste phenotypes ().
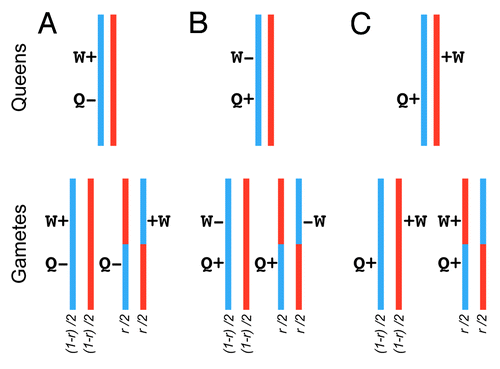
Figure 1. Recombination and the evolution of caste specialization. Recombination can enhance concurrent selection on initially linked mutations that influence queen and worker traits. We assume that both mutations are recent (i.e., have low frequency). We depict frequencies of the different gametes assuming a recombination rate r, which ranges between 0 to 50% (i.e., free recombination). (A) A haplotype containing a slightly deleterious mutation affecting queens linked to a beneficial mutation affecting worker traits and enhancing colony fitness. Recombination can generate a haplotype containing the beneficial worker mutation and the wild-type queen allele, thereby facilitating both positive selection (on the ‘worker’ mutation) and purifying selection (on the ‘queen’ mutation). (B). A haplotype containing a beneficial mutation affecting a queen trait linked to a slightly deleterious mutation affecting a worker trait. Recombination can facilitate concurrent positive selection (on the ‘queen’ mutation) and purifying selection (on the ‘worker’ mutation). (C) Two haplotypes, each containing a beneficial mutation affecting a queen, and a worker trait respectively. These two mutations are on different haplotypes but are spatially proximate. Without recombination, selection will fix one haplotype at the expense of the other. However, one of the recombinant gametes will contain both beneficial mutations allowing for concurrent positive selection on queen and worker traits.
Evolution of Worker Behavior
Workers are the most abundant caste in eusocial insects, and their behavior strongly affects colony fitness.Citation37 By contrasting the behavior and degree of caste specialization between primitively and advanced eusocial insects, it is clearly evident how the evolution of worker behavior and specialization is associated with increased colony sizes and social complexity.Citation38 Given the major contributions of worker behavior to colony fitness,Citation8,Citation39 it is inescapable that selection must be acting on genes that influence worker behavior and enhance colony fitness,Citation40 increasing both linkage disequilibrium and the odds of interference between linked mutations. Indeed, our recent work on a small subset of honey bee genes associated with worker behavior has shown a prevalence of natural selection and high linkage disequilibrium near selective sweeps.Citation41 We have also demonstrated that genes associated with behavior and neurobiology were ancestrally found in regions of the genome with above average GC-content.Citation2 GC-rich areas are expected to contain more mutationsCitation2 and recombination motifs.Citation42 Adaptive evolution of worker behavior is therefore likely to increase linkage disequilibrium and interference selection in GC-rich regions, which in turn can select for the evolution of additional recombination hotspots.Citation42
Indirect Genetic Effects between Workers
Although selection on mutations affecting worker traits is relatively weak because it is indirect (i.e., as it changes the fitness of their reproductive kin),Citation35 social interactions between workers can cause measurable indirect effects on colony fitnessCitation43 leading to social regulatory networks shaped by colony-level selection.Citation44 Indirect phenotypic effects occur when individuals change phenotypes as a result of interactions with others.Citation45 Recent theoryCitation46 suggests that behavioral interaction effects may strongly increase the selection coefficients of mutations influencing interaction traits; this increase in selection coefficients is in theory proportional to the number of interactions among individualsCitation46 and is expected to be greater in larger colonies. If colony fitness increases due to increased worker-worker interactions, selection on mutations with indirect effects on workers can be substantial. Similar to scenario 2, selection on mutations with indirect genetic effects on worker behavior will increase linkage disequilibrium and interference selection, which in turn can select for the evolution of higher recombination rates.
We propose that the evolution of eusociality and recombination rates have been linked in a feedback loop that has strongly affected genome evolution (). Higher recombination rates were selected for during the early stages of the evolution of sociality because they reduce interference selection allowing natural selection to optimize caste phenotypes. This enhanced division of labor then allowed colony sizes to increase, which in turn drives additional reductions in effective population size and concurrent selection for higher recombination rates.
Figure 2. Conceptual model for the evolution of eusociality and recombination rates. Increasing social organization and caste complexity lead to colonies with more workers per queen, reducing effective population size Ne. Lower Ne increases linkage disequilibrium and fixation of mildly deleterious mutations, especially in genomic regions with low recombination. This increases Hill-Robertson interference that can in turn select for higher R, which allows natural selection to act more efficiently on mutations affecting worker traits such as behavior. This in turn facilitates the evolution of higher social complexity and colony sizes that further depress Ne.
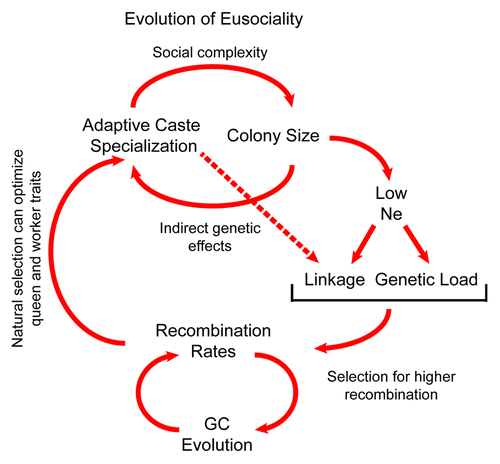
Some components of the loop cause neutral or non-adaptive patterns of molecular and genome evolution, such as the effects of reduced effective population size as colonies grow larger, and the effects of biased gene conversion on GC content. A side effect of increased GC-content due to biased gene conversion may be to increase the number of potential binding sites for fast-evolving recombination hotspot binding factors (such as PRDM9 in mammalsCitation47,Citation48).This phenomenon, in turn, generates new recombination hotspots in GC-rich regions, which replaces recombination hotspots that burn out.Citation48 Our model suggests that not all unusual genomic features found in eusocial insects were caused by adaptive evolutionary forces.
Our model leads to several testable predictions. Our assumption that mutations affecting queens and worker traits are physically proximate—representing one of three possible scenarios that can select for high R—can be tested with more knowledge on the pattern of gene expression, methylation, alternative splicing and gene duplication in sequenced eusocial genomes. Emerging techniques, such as genome wide association mapping, will allow us to link genetic polymorphisms with queen and worker traits in the foreseeable future. Further, population genomic studies will inform us about the prevalence of natural selection and its targets in eusocial organisms. Additionally, our model predicts a negative relationship between recombination rates and GC content with effective population size across social species. Finally, our model predicts some fundamental differences in the degree of linkage disequilibrium, interference selection and the efficiency of positive selection, between the early and late stages of the evolution of sociality. It would be interesting to contrast advanced eusocial and primitively eusocial taxa, and their closely related solitary ancestors, at some of the key population genetic parameters discussed herein. Studies on species that recently reverted from social to solitary lifestylesCitation49 are also relevant, as we believe that these species may not have passed the evolutionary hurdle imposed by interference selection during the initial stages of the evolution of sociality.
We hope our conceptual model stimulates work on genomic and population genomic studies of eusocial taxa, as well as theoretical studies on the extent to which haplodiploidy and eusociality provide the necessary conditions for the evolution of high recombination rates.
Acknowledgments
This study was funded by a Discovery Grant from the Natural Sciences and Engineering Research Council of Canada (NSERC) and an Early Researcher Award from the Ontario Ministry of Research and Innovation (to A.Z.). C.K. was supported by a NSERC Postdoctoral Fellowship. We thank T.A. Linksvayer, L. Packer, B.A.H. Harpur and N. Tsvetkov for helpful comments on the manuscript.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- Wilfert L, Gadau J, Schmid-Hempel P. Variation in genomic recombination rates among animal taxa and the case of social insects. Heredity (Edinb) 2007; 98:189 - 97; http://dx.doi.org/10.1038/sj.hdy.6800950; PMID: 17389895
- Kent CF, Minaei S, Harpur BA, Zayed A. Recombination is associated with the evolution of genome structure and worker behavior in honey bees. Proc Natl Acad Sci USA 2012; 109:18012 - 7; http://dx.doi.org/10.1073/pnas.1208094109; PMID: 23071321
- Duret L, Galtier N. Biased gene conversion and the evolution of mammalian genomic landscapes. Annu Rev Genomics Hum Genet 2009; 10:285 - 311; http://dx.doi.org/10.1146/annurev-genom-082908-150001; PMID: 19630562
- Duret L, Arndt PF. The impact of recombination on nucleotide substitutions in the human genome. PLoS Genet 2008; 4:e1000071; http://dx.doi.org/10.1371/journal.pgen.1000071; PMID: 18464896
- Hodgkinson A, Eyre-Walker A. Variation in the mutation rate across mammalian genomes. Nat Rev Genet 2011; 12:756 - 66; http://dx.doi.org/10.1038/nrg3098; PMID: 21969038
- Gadau J, Helmkampf M, Nygaard S, Roux J, Simola DF, Smith CR, et al. The genomic impact of 100 million years of social evolution in seven ant species. Trends Genet 2012; 28:14 - 21; PMID: 21982512
- Meznar ER, Gadau J, Koeniger N, Rueppell O. Comparative linkage mapping suggests a high recombination rate in all honeybees. J Hered 2010; 101:Suppl 1 S118 - 26; http://dx.doi.org/10.1093/jhered/esq002; PMID: 20212006
- Mattila HR, Seeley TD. Genetic diversity in honey bee colonies enhances productivity and fitness. Science 2007; 317:362 - 4; http://dx.doi.org/10.1126/science.1143046; PMID: 17641199
- Tarpy DR. Genetic diversity within honeybee colonies prevents severe infections and promotes colony growth. Proc Biol Sci 2003; 270:99 - 103; http://dx.doi.org/10.1098/rspb.2002.2199; PMID: 12596763
- Oldroyd BP, Fewell JH. Genetic diversity promotes homeostasis in insect colonies. Trends Ecol Evol 2007; 22:408 - 13; http://dx.doi.org/10.1016/j.tree.2007.06.001; PMID: 17573148
- Sirviö A, Gadau J, Rueppell O, Lamatsch D, Boomsma JJ, Pamilo P, et al. High recombination frequency creates genotypic diversity in colonies of the leaf-cutting ant Acromyrmex echinatior. J Evol Biol 2006; 19:1475 - 85; http://dx.doi.org/10.1111/j.1420-9101.2006.01131.x; PMID: 16910978
- Rueppell O, Meier S, Deutsch R. Multiple mating but not recombination causes quantitative increase in offspring genetic diversity for varying genetic architectures. PLoS ONE 2012; 7:e47220; http://dx.doi.org/10.1371/journal.pone.0047220; PMID: 23077571
- Barton NH. Genetic linkage and natural selection. Philos Trans R Soc Lond B Biol Sci 2010; 365:2559 - 69; http://dx.doi.org/10.1098/rstb.2010.0106; PMID: 20643746
- Barton NH. Mutation and the evolution of recombination. Philos Trans R Soc Lond B Biol Sci 2010; 365:1281 - 94; http://dx.doi.org/10.1098/rstb.2009.0320; PMID: 20308104
- Crozier RH. Counter-intuitive property of effective population size. Nature 1976; 262:384; http://dx.doi.org/10.1038/262384a0; PMID: 958388
- Packer L, Owen R. Population genetic aspects of pollinator decline. Conserv Ecol 2001; 5:4
- Nomura T, Takahashi J. Effective population size in eusocial Hymenoptera with worker-produced males. Heredity (Edinb) 2012; 109:261 - 8; http://dx.doi.org/10.1038/hdy.2012.11; PMID: 22948186
- Chapman RE, Bourke AFG. The influence of sociality on the conservation biology of social insects. Ecol Lett 2001; 4:650 - 62; http://dx.doi.org/10.1046/j.1461-0248.2001.00253.x
- Keightley PD, Otto SP. Interference among deleterious mutations favours sex and recombination in finite populations. Nature 2006; 443:89 - 92; http://dx.doi.org/10.1038/nature05049; PMID: 16957730
- Kimura M. The Neutral Theory of Molecular Evolution. Cambridge, UK: Cambridge University Press, 1985.
- Smith JM, Haigh J. The hitch-hiking effect of a favourable gene. Genet Res 1974; 23:23 - 35; http://dx.doi.org/10.1017/S0016672300014634; PMID: 4407212
- Hill WG, Robertson A. The effect of linkage on limits to artificial selection. Genet Res 1966; 8:269 - 94; http://dx.doi.org/10.1017/S0016672300010156; PMID: 5980116
- Coop G, Przeworski M. An evolutionary view of human recombination. Nat Rev Genet 2007; 8:23 - 34; http://dx.doi.org/10.1038/nrg1947; PMID: 17146469
- Fledel-Alon A, Leffler EM, Guan Y, Stephens M, Coop G, Przeworski M. Variation in human recombination rates and its genetic determinants. PLoS ONE 2011; 6:e20321; http://dx.doi.org/10.1371/journal.pone.0020321; PMID: 21698098
- Hadad R, Pfeiffer T, Poneleit C. Repeatability and heritability of divergent recombination frequencies in the Iowa Stiff Stalk Synthetic (Zea mays L.). TAG. Theor Appl Genet 1996; 93:990 - 6; http://dx.doi.org/10.1007/BF00224103
- Wurm Y, Wang J, Riba-Grognuz O, Corona M, Nygaard S, Hunt BG, et al. The genome of the fire ant Solenopsis invicta. Proc Natl Acad Sci USA 2011; 108:5679 - 84; http://dx.doi.org/10.1073/pnas.1009690108; PMID: 21282665
- Hasselmann M, Lechner S, Schulte C, Beye M. Origin of a function by tandem gene duplication limits the evolutionary capability of its sister copy. Proceedings of the National Academy of Sciences 2010.
- Lyko F, Foret S, Kucharski R, Wolf S, Falckenhayn C, Maleszka R. The honey bee epigenomes: differential methylation of brain DNA in queens and workers. PLoS Biol 2010; 8:e1000506; http://dx.doi.org/10.1371/journal.pbio.1000506; PMID: 21072239
- Toth AL, Varala K, Henshaw MT, Rodriguez-Zas SL, Hudson ME, Robinson GE. Brain transcriptomic analysis in paper wasps identifies genes associated with behaviour across social insect lineages. Proc Biol Sci 2010; 277:2139 - 48; http://dx.doi.org/10.1098/rspb.2010.0090; PMID: 20236980
- Grozinger CM, Fan Y, Hoover SE, Winston ML. Genome-wide analysis reveals differences in brain gene expression patterns associated with caste and reproductive status in honey bees (Apis mellifera). Mol Ecol 2007; 16:4837 - 48; http://dx.doi.org/10.1111/j.1365-294X.2007.03545.x; PMID: 17927707
- Zayed A, Naeger NL, Rodriguez-Zas SL, Robinson GE. Common and novel transcriptional routes to behavioral maturation in worker and male honey bees. Genes Brain Behav 2012; 11:253 - 61; http://dx.doi.org/10.1111/j.1601-183X.2011.00750.x; PMID: 22050787
- Lyko F, Foret S, Kucharski R, Wolf S, Falckenhayn C, Maleszka R. The honey bee epigenomes: differential methylation of brain DNA in queens and workers. PLoS Biol 2010; 8:e1000506; http://dx.doi.org/10.1371/journal.pbio.1000506; PMID: 21072239
- Gadagkar R. The evolution of caste polymorphism in social insects: Genetic release followed by diversifying evolution. J Genet 1997; 76:167 - 79; http://dx.doi.org/10.1007/BF02932215
- Page R, Linksvayer TA, Amdam G. Social life from solitary regulatory networks: a paradigm for insect sociality. Organization of Insect Societies: From Genome to Sociocomplexity. Cambridge, Massachusetts: Harvard University Press, 2009.
- Linksvayer TA, Wade MJ. Genes with social effects are expected to harbor more sequence variation within and between species. Evolution 2009; 63:1685 - 96; http://dx.doi.org/10.1111/j.1558-5646.2009.00670.x; PMID: 19245396
- Hall DW, Goodisman MAD. The effects of kin selection on rates of molecular evolution in social insects. Evolution 2012; 66:2080 - 93; http://dx.doi.org/10.1111/j.1558-5646.2012.01602.x; PMID: 22759286
- Wray MK, Mattila HR, Seeley TD. Collective personalities in honeybee colonies are linked to colony fitness. Anim Behav 2011; 81:559 - 68; http://dx.doi.org/10.1016/j.anbehav.2010.11.027
- Michener CD. The bees of the world. Baltimore, Maryland: The Johns Hopkins University Press, 2000.
- Fewell JH, Winston ML. Colony state and regulation of pollen foraging in the honey bee, Apis mellifera L. Behav Ecol Sociobiol 1992; 30:387 - 93; http://dx.doi.org/10.1007/BF00176173
- Wilson EO. The sociogenesis of insect colonies. Science 1985; 228:1489 - 95; http://dx.doi.org/10.1126/science.228.4707.1489; PMID: 17831241
- Kent CF, Issa A, Bunting AC, Zayed A. Adaptive evolution of a key gene affecting queen and worker traits in the honey bee, Apis mellifera. Mol Ecol 2011; 20:5226 - 35; http://dx.doi.org/10.1111/j.1365-294X.2011.05299.x; PMID: 21981322
- Bessoltane N, Toffano-Nioche C, Solignac M, Mougel F. Fine scale analysis of crossover and non-crossover and detection of recombination sequence motifs in the honeybee (Apis mellifera). PLoS ONE 2012; 7:e36229; http://dx.doi.org/10.1371/journal.pone.0036229; PMID: 22567142
- Linksvayer TA. Direct, maternal, and sibsocial genetic effects on individual and colony traits in an ant. Evolution 2006; 60:2552 - 61; PMID: 17263116
- Linksvayer TA, Fondrk MK, Page RE Jr.. Honeybee social regulatory networks are shaped by colony-level selection. Am Nat 2009; 173:E99 - 107; http://dx.doi.org/10.1086/596527; PMID: 19140771
- Kent C, Azanchi R, Smith B, Formosa A, Levine JD. Social context influences chemical communication in D. melanogaster males. Curr Biol 2008; 18:1384 - 9; http://dx.doi.org/10.1016/j.cub.2008.07.088; PMID: 18789689
- McGlothlin JW, Moore AJ, Wolf JB, Brodie ED 3rd. Interacting phenotypes and the evolutionary process. III. Social evolution. Evolution 2010; 64:2558 - 74; http://dx.doi.org/10.1111/j.1558-5646.2010.01012.x; PMID: 20394666
- Ponting CP. What are the genomic drivers of the rapid evolution of PRDM9?. Trends Genet 2011; 27:165 - 71; http://dx.doi.org/10.1016/j.tig.2011.02.001; PMID: 21388701
- Myers S, Bowden R, Tumian A, Bontrop RE, Freeman C, MacFie TS, et al. Drive against hotspot motifs in primates implicates the PRDM9 gene in meiotic recombination. Science 2010; 327:876 - 9; http://dx.doi.org/10.1126/science.1182363; PMID: 20044541
- Danforth BN, Conway L, Ji S. Phylogeny of eusocial Lasioglossum reveals multiple losses of eusociality within a primitively eusocial clade of bees (Hymenoptera: Halictidae). Syst Biol 2003; 52:23 - 36; http://dx.doi.org/10.1080/10635150390132687; PMID: 12554437