Abstract
Molluscs are a large and diverse group of aquatic and terrestrial animals that rely heavily on chemical communication. Aplysia is an excellent model in which to investigate and develop breakthrough principles into the molecular aspects of chemoreception in molluscs. We recently identified a large family of rhodopsin-like G-protein coupled receptors expressed in the chemosensory rhinophore of Aplysia that may be key components of sensory detection. Here, we summarize these findings and provide further insight into the molecular olfactory toolkit used by Aplysia, by taking advantage of our knowledge of their attraction pheromones. Our characterization of rhinophore genes upregulated following pheromone stimulation helps explain the dynamics of olfactory gene expression following chemical stimulation.
Chemical signals are known to convey a remarkable amount of information. To understand how this occurs, research has been devoted towards identifying the compounds that are involved, how they are produced, and how olfactory organs are able to ‘decode’ information from chemical signals. Although well studied in the broad behavioral and physiological context, there is relatively little knowledge of how olfaction works at the molecular level in a huge diversity of aquatic invertebrate animals, from corals and sponges to octopus and oysters. Incredibly, despite molluscs representing the second most abundant living phyla and the most diverse,Citation1 it is only now that we are starting to elucidate the molecular toolkit used by molluscs to enable them to smell.
The seminal olfactory discoveries obtained at the molecular and cellular level from the mouse (representing chordates), Drosophila (arthropods) and Caenorhabditis (nematodes) provides compelling evidence for the necessity to study scientifically tractable animals, prior to exploration into those of commercial relevance. For example, the identification of olfactory receptors in Drosophila in 1999,Citation2 was pivotal to the research that led to the multitude of olfactory-related entomological discoveries within the insect group, including the moth, honeybee and mosquito. We need to interrogate the molluscan olfactory system in a similar fashion.
The first stage of olfaction is handled by the sensory neurons in the olfactory epithelium where they express genes that encode products devoted to binding environmental odorants and transferring this information intracellular. In mammals, detection of environmental chemicals results from binding of molecules to one or more of a large family of seven-transmembrane domain rhodopsin and metabotropic G-protein coupled receptors (GPCRs) that are expressed almost exclusively on the surface of sensory neurons within the main olfactory epithelium and vomeronasal organ.Citation3 Meanwhile, Benton et al. 2009,Citation4 were first to discover that variant ionotropic receptors were chemosensory in insects and appear to enhance processing speed. Insect olfactory systems respond to a smaller range of odorants than terrestrial vertebrates, and their genomes in turn encode a smaller family of receptors.Citation5
We use Aplysia as our model system as it is almost unique amongst molluscs in that it has been highly characterized at multiple levels and therefore fulfils many of the requirements necessary to investigate the molecular basis of olfaction. First, various aspects of the structure and function of olfactory organs, life history, such as larval development and laboratory rearing, have been well described.Citation6,Citation7 Second, the availability of multiple expressed sequence tag transcriptome libraries, including central neuron transcripts,Citation8 as well as the Aplysia genome, enables detailed genetic analyses. Finally and perhaps most important, they have no acoustic sense and little to no ability to recognise objects by vision, instead, their world is chemically driven and many key physiological and behavioral events must be mediated by secreted water-soluble chemicals, leading to aggregation, habitat selection, defence and courtship and mating. For example, induction of larval metamorphosis requires a chemical cue from algal sources.Citation9 Also, mate attraction and subsequent mating is stimulated by the release of conspecific water-borne sexual pheromones.Citation10 Experimental attraction assays followed by molecular and biochemical analysis were important to demonstrate that Aplysia had developed a potent solution to attracting a mate by releasing attraction pheromones, consisting of four small proteins that were subsequently named attractin, enticin, temptin and seductinCitation11 ().
At the anatomical level, odorant detection is achieved by a pair of rhinophore, specialized anterior sensory organs on the dorsal surface of the head that act as a finely tuned nose.Citation7 These also allow animals to locate and discriminate food, conspecific pheromones and predators. From the rhinophore sensory epithelium we identified genes encoding proteins that may directly or indirectly be involved in Aplysia olfaction, including, a large multi-gene receptor family encoding rhodopsin-like GPCRs (>90), identified by analyzing the unfinished Aplysia genome database and using algorithms that identify genes encoding multi-transmembrane proteins.Citation12 Phylogenetic analysis revealed they consist of three distinct monophyletic protein subfamilies and expression analysis using laser capture microdissection-reverse transcription-polymerase chain reaction and sequencing revealed expression of at least some of these genes within the sensory neurons located within the rhinophore groove. These gene families encode novel candidate chemoreceptors that show only distant identity with known vertebrate, insect and nematode chemoreceptors.Citation12,Citation13 Based on molecular genetic and expression analyses, Aplysia G proteins, an IP3 receptor, and a phopholipase C, are expressed and localise to the olfactory sensory epithelium of the rhinophore.Citation14 This suggests that the receptors identified may be G-protein coupled, similar to that of vertebrates. However, Aplyisa also appear to have their own variant ionotropic receptors, similar to that found in insects, some of which are found in rhinophore. (V. Croset, S. Cummins, R. Benton, in preparation) Further understanding of these will significantly expand our understanding of how these animals detect water-soluble chemicals.
Our knowledge of the attraction pheromones and site of pheromone detection provides an excellent opportunity to gain insight into the dynamics of olfactory gene expression. Towards achieving this, we performed a suppressive subtraction hybridization, to identify those genes upregulated following pheromone exposure (method summarized in ). In total, 40 clones were sequenced and identification of unigenes achieved by blastn or tblastn of the NCBI database, resulting in 27 significant (cut-off of 10−6 E-value) matches. These included complementary DNAs (cDNAs) encoding an isoform of a-tubulin and actin, the neuropeptide schistosomin, an Aplysia temptin-like peptide, an inhibition of apoptosis protein and several cDNAs encoding products involved in translation, including ribosomal protein L18a (). The identified temptin-like gene shows similarity with the previously described temptin pheromoneCitation15 only within the conserved calcium-binding EGF-like domain. The cytoskeletal α1-tubulin is known to be present not only in the cell bodies but also in the distal neurites of cultured Aplysia californica sensory neurons, where serotonin can induce its specific translation.Citation16 Thirteen unigenes remain unidentified. Of those, two cDNAs contain a 303 base pair open reading frame that encode a 91 amino acid precursor predicted to be post-translationally cleaved and secreted extracellular (). A similar gene has also been identified from the Aplysia kurodai neural transcriptomeCitation17 (), suggesting a neuronal origin and conservation of function. All of the genes identified from this subtraction may be involved in pheromone detection, processing or modulation.
The identification of these, along with the candidate chemoreceptors,Citation12 will enable their use as tools to further investigate the dynamics of olfactory gene expression and provide a critical bridge between olfactory genes, circuits and behaviour in the broad evolutionary context. As a gastropod mollusc, Aplysia does share many biological characteristics with economically important aquaculture molluscs, including abalone, as well as pest molluscs such as Pomacea, Cernuella, Theba (all crop pests), and Biomphalaria (intermediate parasite host). Indeed, further understanding of the genetic basis of smell in these animals using olfactory principles derived from the Aplysia work may provide opportunities to develop agents that could enhance or disrupt chemical signaling.
Methods
Aplysia dactylomela (12 animals collected from Caloundra, Queensland) were divided into three groups containing four animals each and kept in individual cages containing 2 L filtered sea water. Treatments included: Group (1) Animals were injected with egg-laying hormone (40 µg/ml).Citation18 Sixty minutes later, all egg cordons were transferred to 40 ml fresh filtered sea water and gently shaken for 30 min. Egg eluate (containing pheromones) was then taken for; Group (2) Animals were exposed to 10 ml of freshly collected egg eluate. At 15 min, rhinophore were removed from the base, combined and immediately frozen on dry ice. These comprised pheromone-stimulated rhinophore (PSR). Group (3) Animals were exposed to 10 ml of filtered sea water. At 15 min, rhinophore were removed from the base, combined and immediately frozen on dry ice. These comprised non-stimulated rhinophore (NSR). RNA was isolated from both PSR and NSR with the Oligotex Direct mRNA Mini Kit (Qiagen) as per manufacturer's protocol, and quantitated at 280 nm with a NanoDrop ND-1000 spectrophotometer (NanoDrop Products). Complementary DNA (cDNA) synthesis was performed followed by amplification and suppression subtractive hybridization using the Clontech Super SMART™ PCR cDNA Synthesis Kit (Clontech) and PCR-Select™ cDNA Subtraction Kit (Clontech), respectively. PSR cDNA was used as tester and NSR cDNA as driver. Unidirectional sequencing was performed by the Australian Genome Research Facility. Resulting sequences were vector trimmed and adaptor sequences removed by the CodonCode program (www.codoncode.com), and were checked by manual inspection.
Figures and Tables
Figure 1 Aplysia use water-soluble pheromones to attract potential mating partners. The figure shows Aplysia brasiliana and a model of the pheromones attractin and temptin in complex. Modified from reference Citation19.
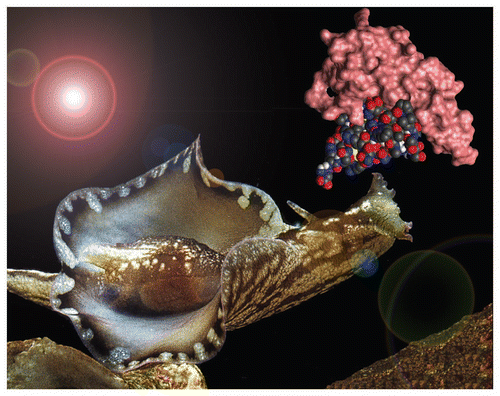
Figure 2 Identification of Aplysia rhinophore genes upregulated following exposure to attraction pheromones. (A) Summary of experimental procedure for gene identification. (B) Nucleotide and predicted amino acid sequences of a novel gene identified (clones 16 and 31), encoding a predicted secreted peptide. An arrow denotes the predicted signal sequence cleavage site, a dibasic cleavage site is underlined (K57R) and cysteines are highlighted in grey. A cartoon schematic representation is shown below. Met, Methionine; Signal, secretion signal sequence. (C) Comparative amino acid alignment of Aplysia dactylomela (Genbank: HM191483) and Aplysia kurodai (Genbank: EY420369) novel gene product. Amino acids that are identical are shaded in black.
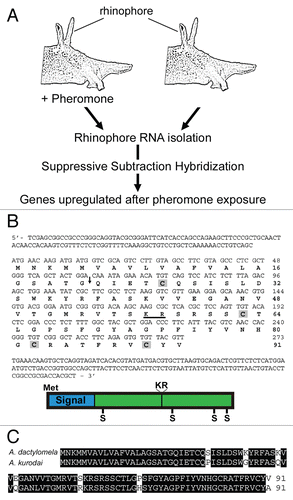
Table 1 Pheromone-stimulated rhinophore/non-stimulated rhinophore subtraction characterized unigenes
Acknowledgements
This work was supported by grants from the Australian Research Council to Bernard Degnan. Scott Cummins was supported by a University of Queensland Fellowship.
Addendum to:
References
- Wagele H, Klussmann-Kolb A. Opisthobranchia (Mollusca, Gastropoda)—more than just slimy slugs. Shell reduction and its implications on defence and foraging. Front Zool 2005; 2:3 - 20
- Clyne PJ, CG W, Freeman MR, Lessing D, Kim J, Carlson JR. A novel family of divergent seven-transmembrane proteins: candidate odorant receptors in Drosophila. Neuron 1999; 22:327 - 338
- Matsunami H, Buck LB. A multigene family encoding a diverse array of putative pheromone receptors in mammals. Cell 1997; 90:775 - 784
- Benton R, Vannice KS, Gomez-Diaz C, Vosshall LB. Variant ionotropic glutamate receptors as chemosensory receptors in Drosophila. Cell 2009; 136:149 - 162
- de Bruyne M, Warr GC. Molecular and cellular organization of insect chemosensory neurons. Bioessays 2006; 28:23 - 24
- Gerdes R, Fieber LA. Life history and aging of captive-reared California sea hares (Aplysia californica). Am Ass Lab Animal Sci 2006; 45:40 - 47
- Wertz A, Rössler W, Obermayer M, Bickmeyer U. Functional neuroanatomy of the rhinophore of Aplysia punctata. Front Zool 2006; 3:6
- Moroz LL, Edwards JR, Puthanveettil SV, Kohn AB, Ha T, Heyland A, et al. Neuronal transcriptome of Aplysia: neuronal compartments and circuitry. Cell 2006; 29:1453 - 1467
- Plaut I, Borut A, Spira ME. Growth and metamorphosis of Aplysia oculifera larvae in laboratory culture. Marine Biol 1995; 122:425 - 430
- Painter SD, Cummins SF, Nichols AE, Akalal DB, t Schein CH, Braun W, et al. Structural and functional analysis of Aplysia attractins, a family of water-borne protein pheromones with interspecific attractiveness. Proc Natl Acad Sci USA 2004; 101:6929 - 6933
- Cummins SF, Nichols AE, Amare A, Hummon AB, Sweedler JV, Nagle GT. Characterization of Aplysia enticin and temptin, two novel water-borne protein pheromones that act in concert with attractin to stimulate mate attraction. J Biol Chem 2004; 279:25614 - 25622
- Cummins SF, Erpenbeck D, Zou Z, Claudianos C, Moroz LL, Nagle GT, et al. Candidate chemoreceptor subfamilies differentially expressed in the chemosensory organs of the mollusc. BMC Biol 2009; 7
- Cummins SF, LeBlanc L, Degnan BM, Nagle GT. Molecular identification of candidate chemoreceptor genes and signal transduction components in the sensory epithelium of Aplysia. J Exp Biol 2009; 212:2037 - 2044
- Cummins SF, De Vries MR, Hill KS, Boehning D, Nagle GT. Gene identification and evidence for expression of G protein alpha subunits, phospholipase C, and an inositol 1,4,5-trisphosphate receptor in Aplysia californica rhinophore. Genomics 2007; 90:110 - 120
- Cummins SF, Xie F, de Vries MR, Annangudi SP, Misra M, Degnan BM, et al. Aplysia temptin—the ‘glue’ in the water-borne attractin pheromone complex. FEBS J 2007; 274:5425 - 5437
- Moccia R, Chen D, Lyles V, Kapuya E, Kalachikov S, Spahn CM, et al. An unbiased cDNA library prepared from isolated Aplysia sensory neuron processes is enriched for cytoskeletal and translational mRNAs. J Neurosci 2003; 23:9409 - 9417
- Lee YS, Choi SL, Kim TH, Lee JA, Kim HK, Kim H, et al. Transcriptome analysis and identification of regulators for long-term plasticity in Aplysia kurodai. Proc Natl Acad Sci USA 2008; 105:18602 - 18607
- Cummins SF, Nuurai P, Nagle GT, Degnan BM. Conservation of the egg-laying hormone neuropeptide and attractin pheromone in the spotted sea hare, Aplysia dactylomela. Peptides 2010; 31:394 - 401
- Cummins SF, Xie F, de Vries MR, Annangudi SP, Misra M, Degnan BM, et al. Aplysia temptin—the ‘glue’ in the water-borne attractin pheromone complex. FEBS J 2007; 274:5425 - 5437