Abstract
Formation of a new memory requires plasticity at the synaptic level. However, it has also been shown that the consolidation and the maintenance of such a new memory involve processes that necessitate active mRNA at the nucleus of the cell. How can robust changes in synaptic efficacy specifically drive new transcription and translation of new gene transcripts, and thus transform an otherwise transient plasticity into a long-lasting and stable one? In this article, we highlight the conceptual advance that was gained by the discovery of a potent Synaptic Activity Responsive Element (SARE) found ~7 kb upstream of the transcription initiation site of the neuronal immediate early gene Arc. The unique genomic structure of SARE, which contained adjacent and cooperative binding sites for three major activity-dependent transcription factors within a 100-bp locus, was associated with an unusual responsiveness to neuronal stimuli. Taken together, these findings shed light on a new class of transcriptional sensor with enhanced sensitivity to synaptic activity.
Strengthening and weakening of synaptic connectivity have been thought to underlie the plastic changes that occur within neuronal circuits, when an animal has once experienced a robust environmental change such that its context and content have to be recalled over time for this animal to survive.Citation1 Protein synthesis inhibitors have been shown to block such storage of new information. One influential hypothesis in the field of learning and memory (termed “the synaptic tagging and capture hypothesis”)Citation2 postulated that formation of a robust memory may trigger plasticity at stimulated synapses, while also may strongly induce expression of specific sets of genes in the nucleus. The subsequent translation and the trafficking of these newly transcribed genes into the plastic “tagged” synapses appeared to provide an attractive mechanism that could successfully account for the segregation of stably plastic synapses (in which “tags” were able to functionally “capture” new gene products) from weakly plastic ones (which could not capture any).
What could be the transcriptional regulators that are activated in conjunction with synaptic plasticity? Are there many of them? One of the obvious candidates is the cyclic AMP-responsive element binding protein (CREB), a transcription factor that has been shown to be involved in long-term memory formation, consolidation and reconsolidation.Citation3–Citation8 Adult mice with disrupted CREB function in the brain exhibited a profound and specific impairment in long-term memory while the short-term memory remained unaltered.Citation3,Citation5,Citation6 Consistent with a potential role of CREB, several intracellular signaling pathways that are stimulated by synaptic activity, such as CaMKK-CaMKIV cascade, cAMP/PKA stimulation or Ras/Raf/MEK/ERK cascade have also been shown to elevate levels of CREB phosphorylation status.Citation9–Citation13
However, more than 5% of mammalian genes appear to be potentially regulated by CREB-dependent transcription, as determined by genome-wide analyses of CREB-bound promoters.Citation14,Citation15 How can we then pinpoint the major CREB target genes that are involved in long-term memory maintenance? For example, c-fos or brain-derived neurotrophic factor (BDNF) genes possess well-known consensus CRE elements in their proximal promoter regions.Citation16–Citation18 In sharp contrast, although many recent studies had highlighted the critical importance of Activity-regulated cytoskeleton-associated protein (Arc) gene,Citation19–Citation25 both as an accurate and sensitive marker for enhanced cognitive activity,Citation26,Citation27 and as a memory-forming gene,Citation23,Citation28 no functional CRE site was reported.Citation29 Was Arc induced by synaptic activity by a mechanism that did not require CREB? Or were there hidden CRE sites that needed to be revealed?
Kawashima et al.Citation30 employed an ameliorated promoter assay system in cortical neuronal cultures that was optimized for synaptic stimulation and gene transfer of large plasmids. Through careful promoter analyses, the critical responsible element was pinned down to about 100-bp, which was named Synaptic Activity-Responsive Element (SARE). This element, when placed in isolation next to a minimal promoter region, could still trigger an extremely high level of gene induction upon receipt of synaptic activity, to an extent comparable to the full-length ∼7-kb promoter (). By scrutinizing the SARE sequence, and through a combination of electrophoretic mobility shift and chromatin immunoprecipitation assays, it became evident that SARE had a unique structure consisting of a half CRE site that was juxtaposed to a MEF2 site and an SRF/TCF site. Point mutations in either one of these sites potently attenuated the synaptic activity-dependency, indicating that the CREB-, MEF2- and SRF-binding sites within SARE needed to be co-occupied in order to reach full potency. Pharmacological experiments determined the requirement for both CaMK- and MAPK-dependent pathways in this activation process ().
This study has shed light on the new exciting possibility that the co-occupancy of 3 major activity-dependent transcription factor sites in close proximity within SARE is not only necessary but also sufficient to trigger an unusually large synaptic activity-induced transcriptional response. Future studies are clearly needed to elucidate how neuronal activity can regulate the occupancy of individual sites, and determine the sustainability of this putative transcriptional complex. Finally, the potential role of Arc as a gene product captured and working at “tagged” synapses still needs intense investigation. Further deciphering of the signaling from synapse to the nucleus and back to the original stimulated synapses will hopefully pave the way for better understanding of cognitive disorders including mental retardation and memory deficits.
Figures and Tables
Figure 1 Synaptic Activity-Responsive Element (SARE) possesses a strong enhancer activity that is uniquely sensitive in response to synaptic stimulation. (A) SARE locates in an evolutionarily conserved genomic region in the Arc promoter. The SARE transcriptional activity was investigated with a construct in which SARE was fused directly upstream of a short TATA-containing sequences of the Arc promoter (SARE-ArcTATA). (B) SARE replicates the activation ability of the Arc7000 full promoter. Left, Summary of luciferase assays. Cultured cortical neurons electroporated with luciferase reporter constructs were synaptically activated and luciferase induction-folds were measured. Right, GFP reporter assays. GFP reporter expression via SARE activation was detected in individual neurons. RFP served as a transfection marker. Modified from Kawashima et al.Citation30 Scale bar, 50 µm.
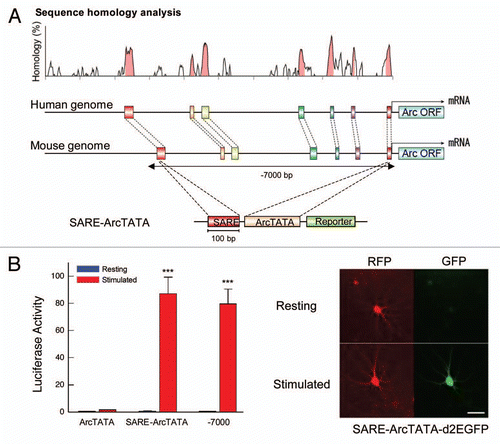
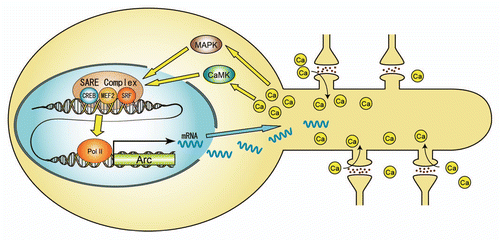
Figure 2 A model of activity-dependent regulation of the Arc gene. Ca2+ entry through NMDA-receptors activates CaMK- and MAPK-pathways, which in turn activate 3 major acivitity-responsive transcription factors, CREB, MEF2 and SRF at the SARE site. A putative SARE-protein complex interacts with the transcriptional machinery to promote Arc gene epxression.
Acknowledgements
We apologize to the authors whose contributions could not be cited due to space limitations. This work was supported in part by grants-in-aid from the Ministry of Education, Culture, Sports, Science and Technology of Japan (M.N., H.O., H.B.) and the Ministry of Health, Labour and Welfare of Japan (H.B.), and by an award from the Astellas Foundation for Research on Metabolic Disorders (H.O.). T.K. was a predoctoral fellow funded by Japan Society for the Promotion of Science.
Addendum to:
References
- Bliss TV, Collingridge GL. A synaptic model of memory: long-term potentiation in the hippocampus. Nature 1993; 361:31 - 39
- Morris RG. Elements of a neurobiological theory of hippocampal function: the role of synaptic plasticity, synaptic tagging and schemas. Eur J Neurosci 2006; 23:2829 - 2846
- Bourtchuladze R, et al. Deficient long-term memory in mice with a targeted mutation of the cAMP-responsive element-binding protein. Cell 1994; 79:59 - 68
- Bito H, Deisseroth K, Tsien RW. CREB phosphorylation and dephosphorylation: a Ca(2+)- and stimulus duration-dependent switch for hippocampal gene expression. Cell 1996; 87:1203 - 1214
- Silva AJ, et al. CREB and memory. Annu Rev Neurosci 1998; 21:127 - 148
- Kida S, et al. CREB required for the stability of new and reactivated fear memories. Nat Neurosci 2002; 5:348 - 355
- Conkright MD, Montminy M. CREB: the unindicted cancer co-conspirator. Trends Cell Biol 2005; 15:457 - 459
- Nonaka M. A Janus-like role of CREB protein: enhancement of synaptic property in mature neurons and suppression of synaptogenesis and reduced network synchrony in early development. J Neurosci 2009; 29:6389 - 6391
- Gonzalez GA, Montminy MR. Cyclic AMP stimulates somatostatin gene transcription by phosphorylation of CREB at serine 133. Cell 1989; 59:675 - 680
- Ginty DD, Bonni A, Greenberg ME. Nerve growth factor activates a Ras-dependent protein kinase that stimulates c-fos transcription via phosphorylation of CREB. Cell 1994; 77:713 - 725
- Bito H, Deisseroth K, Tsien RW. Ca2+-dependent regulation in neuronal gene expression. Curr Opin Neurobiol 1997; 7:419 - 429
- Shaywitz AJ, Greenberg ME. CREB: a stimulus-induced transcription factor activated by a diverse array of extracellular signals. Annu Rev Biochem 1999; 68:821 - 861
- Bito H, Takemoto-Kimura S. Ca2+/CREB/CBP-dependent gene regulation: a shared mechanism critical in long-term synaptic plasticity and neuronal survival. Cell Calcium 2003; 34:425 - 430
- Impey S, et al. Defining the CREB regulon: a genome-wide analysis of transcription factor regulatory regions. Cell 2004; 119:1041 - 1054
- Zhang X, et al. Genome-wide analysis of cAMP-response element binding protein occupancy, phosphorylation, and target gene activation in human tissues. Proc Natl Acad Sci USA 2005; 102:4459 - 4464
- Robertson LM, et al. Regulation of c-fos expression in transgenic mice requires multiple interdependent transcription control elements. Neuron 1995; 14:241 - 252
- West AE, Griffith EC, Greenberg ME. Regulation of transcription factors by neuronal activity. Nat Rev Neurosci 2002; 3:921 - 931
- Flavell SW, Greenberg ME. Signaling mechanisms linking neuronal activity to gene expression and plasticity of the nervous system. Annu Rev Neurosci 2008; 31:563 - 590
- Link W, et al. Somatodendritic expression of an immediate early gene is regulated by synaptic activity. Proc Natl Acad Sci USA 1995; 92:5734 - 578
- Lyford GL, et al. Arc, a growth factor and activityregulated gene, encodes a novel cytoskeleton-associated protein that is enriched in neuronal dendrites. Neuron 1995; 14:433 - 445
- Tagawa Y, et al. Multiple periods of functional ocular dominance plasticity in mouse visual cortex. Nat Neurosci 2005; 8:380 - 388
- Chowdhury S, et al. Arc/Arg3.1 interacts with the endocytic machinery to regulate AMPA receptor trafficking. Neuron 2006; 52:445 - 459
- Plath N, et al. Arc/Arg3.1 is essential for the consolidation of synaptic plasticity and memories. Neuron 2006; 52:437 - 444
- Shepherd JD, et al. Arc/Arg3.1 mediates homeostatic synaptic scaling of AMPA receptors. Neuron 2006; 52:475 - 484
- McCurry CL, et al. Loss of Arc renders the visual cortex impervious to the effects of sensory experience or deprivation. Nat Neurosci 2010; 13:450 - 457
- Guzowski JF, et al. Environment-specific expression of the immediate-early gene Arc in hippocampal neuronal ensembles. Nat Neurosci 1999; 2:1120 - 1124
- Ramirez-Amaya V, et al. Spatial exploration-induced Arc mRNA and protein expression: evidence for selective, network-specific reactivation. J Neurosci 2005; 25:1761 - 1768
- Han JH, et al. Neuronal competition and selection during memory formation. Science 2007; 316:457 - 460
- Waltereit R, et al. Arg3.1/Arc mRNA induction by Ca2+ and cAMP requires protein kinase A and mitogen-activated protein kinase/extracellular regulated kinase activation. J Neurosci 2001; 21:5484 - 5493
- Kawashima T, et al. Synaptic activity-responsive element in the Arc/Arg3.1 promoter essential for synapse-to-nucleus signaling in activated neurons. Proc Natl Acad Sci USA 2009; 106:316 - 321