Abstract
The behaviour of a cell is determined by the interplay of its subcellular components. Thus, being able to simultaneously visualize several organelles inside cells within the natural context of a living organism could greatly enhance our understanding of developmental processes. We have established a Gal4-based system for the simultaneous and cell type specific expression of multiple subcellular labels in transparent zebrafish embryos. This system offers the opportunity to follow intracellular developmental processes in a live vertebrate organism using confocal fluorescence time-lapse microscopy. Using this approach we recently showed that the centrosome neither persistently leads migration nor determines the site of axonogenesis in migrating neurons in the zebrafish cerebellum in vivo. Here we present additional in vivo findings about the centrosomal and microtubule dynamics of neuroepithelial cells during mitotic cleavages at early neural tube stages.
The developmental events that shape an embryo involve complex interactions between cells, tissues and extracellular matrix, and so are ideally investigated within the context of a living organism.Citation1 Underlying many morphological changes are the movements of individual cells, which themselves are driven by the dynamics of subcellular organelles. Thus, what happens intracellularly dictates the temporal and spatial orchestration of proliferation, apoptosis, migration, differentiation and morphological specialization, eventually leading to functional organs like the brain.
In cells initiating mitosis, the microtubule cytoskeleton undergoes major remodeling and the centrosomes relocate within the cell to form the spindle poles of the microtubule-based spindle apparatus. Next, replicated chromosomes condense, the nuclear envelope breaks down and microtubules attach to kinetochores to pull sister chromatids apart. At the end of mitosis, cytokinesis separates the two daughter cells. Here, a contractile actomyosin ring drives constriction of the plasma membrane to form a narrow cytoplasmic bridge between the two reforming nuclei. Physical separation of the two daughter cells requires this bridge to be severed in a final step termed abscission. This cytoplasmic bridge consists of a thin tube of plasma membrane filled with two antiparallel bundles of microtubules with their plus-ends interdigitating at the midbody. Live imaging of cells in culture reveals that the bridge can persist for several hours before abscission occurs, through a mechanism that remains largely unknown.Citation2 A current model suggests that the two centrioles separate and the mother centriole translocates to the intercellular bridge, bringing proteins located at the centrosome together with their interaction partners located at the midbody, hereby initiating abscission.Citation3–Citation6
The ability to visualize organelles inside cells within a living organism could greatly enhance our understanding of developmental events such as mitosis. Towards this goal, we have established a system for the expression of multiple subcellular labels in specific cells of a living zebrafish embryo. Owing to their transparency during embryonic stages and their fast and external development, zebrafish are an ideal model organism for in vivo microscopy.Citation7,Citation8 Our labeling technique is based on the Gal4-UAS system, in which Gal4 is a yeast-derived transcriptional activator that induces gene expression by binding to DNA binding sites called upstream activating sequences (UAS).
We have established so-called Medusa vectors (), which contain UAS sites bidirectionally flanked by basal promoters driving expression of subcellularly targeted fluorescent proteins with distinct spectral properties. Thus, multiple organelles can be simultaneously demarcated for confocal fluorescence microscopy. Moreover, by placing Gal4 expression under tissue specific control, as already achieved in many stable transgenic Gal4-driver lines, the expression of organelle labels can be restricted to cell types of interest.Citation9–Citation11
We recently performed Medusa vector labeling of the centrosome, nucleus and membrane in migrating tegmental hind-brain nuclei neurons in the live zebrafish cerebellum, and demonstrated that the centrosome and nucleus exhibit an unexpected sort of leapfrogging behavior during nucleokinesis. Further, we found that the centrosome does not by its proximity determine the site of axonogenesis in these neurons.Citation12
Here we have applied our multiorganelle labeling approach by coinjecting a Gal4 expression vector (pCSKalTA4)together with Medusa M1, M2 or M3 constructsCitation12 at the one cell stage and following by in vivo time-lapse recording the interplay of plasma membrane, nucleus, microtubules and centrosome in neuroepithelial cells, which divide at the midline in the zebrafish hindbrain and anterior spinal cord at early neural tube stages (Sup. Vid. 1).Citation13–Citation16 The two centrioles of the centrosome are located at the apical membrane of neuroepithelial cells.Citation12 We observed, as expected, that the centrosome replicated prior to cell division and the two centrosomes left the apical membrane to form the spindle poles of the spindle apparatus (). Surprisingly though, cleavage furrow ingression did not occur symmetrically, as reported for HeLa cells, but instead progressed almost entirely from the basal side of dividing neuroepithelial cellsCitation17 (–E white arrow and Sup. Vids. 1–3). The asymmetric ingression caused the microtubules of the spindle apparatus to bend towards the midline ( at 840 sec, and D Sup. Vids. 1 and 2 at 840 sec, and D, Sup. Vids. 1 and 2), resulting in an apically localized midbody connecting the two daughter cells ( at 1.120 sec). In addition, our microtubule label condensed lateral to the furrow at late furrow ingression stages (–E). After separation of the chromosomes during cytokinesis, the spindle poles translocated along the plasma membrane towards the apical membrane (–F and Sup. Vids. 2 and 3). The centrosome was located at the spindle pole and visualization of the centrosome by Medusa vector M1 expression revealed an apical movement similar to that seen for the microtubule spindle pole (Sup. Vid. 4). Interestingly, we found that during apically-directed centrosome movements, microtubules still polymerized from the centrosome, mainly lateral to the furrow (Sup. Vid. 3) and the microtubule network of the remnant spindle apparatus remained intact during this relocation. Once the centrosome reached the apical membrane microtubule polymerization continued around the nucleus (Sup. Vid. 3). The rotation and the spatial polymerization dynamics of the microtubule network are likely to be the driving force behind the shape change from round during mitotic phases to apico-basally elongated once the daughter cells have formed (–F). In contrast to the observed splitting of centrioles in HeLa cells, with the mother centriole moving towards the midbody to initiate abscission,Citation3 we found that both centrioles of the centrosome stayed together while relocating to the apical side ( and D and Sup. Vid. 4). These findings indicate the need for further in vivo cell biological studies because, although molecular players have been identified,Citation4,Citation6,Citation18 the mechanisms underlying cytokinesis and abscission are not fully understood and the role of the centrioles in abscission remains controversial.Citation19 Observed discrepancies might be due to cell type differences or differences between the in vitro and in vivo situation. Notably, the versatility of our subcellular labeling system will allow for the comparison of cell biological mechanisms across various cell types.
In addition to labeling subcellular structures, our Gal4-based system can be used for gain- or loss-of-function analyses through the addition of dominant-negative, wildtype or otherwise mutated genes to the Medusa vectors. Thus, the function of candidate genes involved in cytokinesis or abscission can be studied with a direct read-out of the subcellular changes in the context of a differentiating tissue.
Our system has proven to be a powerful tool for the study of developmental processes on a subcellular level in a living organism.Citation12 Our initial applications of these in vivo cell biology tools have already revealed significant differences from current models, promising that there is much new and exciting to discover deep within a zebrafish.
Figures and Tables
Figure 1 Gal4 mediated subcellular labeling. (A) Schematic showing subcellular labeling strategy. A Medusa vector was constructed containing subcellular labels for the nucleus (H2B-mRFP, red), plasma membrane (memCFP-blue) and microtubules (GFP-DCX, green) all under the control of Gal4 DNA binding sites (UAS). By flanking UAS sites with E1b minimal promoters and two expression cassettes (here H2B-mRFP and GFP-DCX), bidirectional expression can be achieved upon Gal4 binding. Tissue specificity can be achieved by expressing Gal4 under control of tissue-specific regulatory elements. (B) Dorsal view of a zebrafish embryo at neural rod to neural tube stages. Anterior is to the left. The red box depicts the area shown in (C) and in and . (C) Neuroepthelial cells expressing Medusa vector M2 depicted in (A). Mitotic cleavages can be observed at the midline (white arrows). At 1120 s, daughter cells connected by a midbody can still be seen.
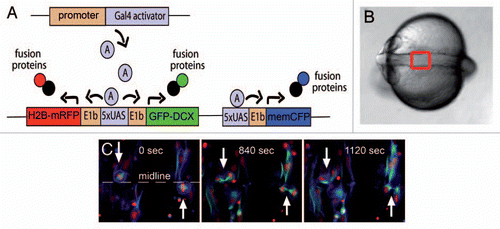
Figure 2 Cytokinesis. Dorsal views of an early neural tube stage zebrafish embryo injected with pCSKalTA 4 and Medusa vector M2. (A) Neuroepithelial cell dividing at the midline. (B) Furrow ingression occurs from the basal side (white arrows in B-D). (C and D) The microtubules of the spindle apparatus bend towards the apical side. (E and F) The spindle pole (white arrow), carrying an intact microtubule cytoskeleton, moves along the plasma membrane towards the forming midbody, thereby changing the shape of the cell from round to apico-basally elongated.
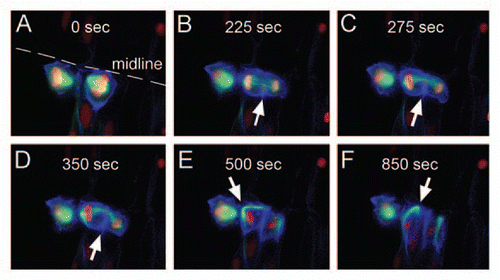
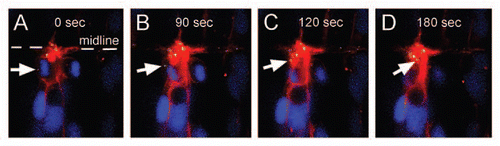
Figure 3 Centrosomal movements during cytokinesis. Dorsal views of an early neural tube stage zebrafish embryo expressing Medusa vector M1, which contains Centrin2-YFP (yellow centrosome), memmRFP (red membrane) and H2B-CFP (blue nucleus). After separation of the sister chromatids, the centrosome relocates from the spindle pole to the apical membrane. The centrioles of the centrosome (white arrow) remain together as they move towards the midbody of neuroepithelial cells.
Additional material
Download Zip (3.9 MB)Acknowledgements
We are grateful for excellent fish husbandry and technical assistance by Petra Hammerl, Enrico Kühn and Christiane Lach. This work was generously supported by the Deutsche Forschungsgemeinschaft (DFG, KO1949/3-1), a Fellowship of the Studienstiftung des deutschen Volkes (M.D.) and a Postdoctoral Fellowship from the Natural Sciences and Engineering Research Council (NSERC) of Canada (J.H.).
Addendum to:
References
- Lichtman JW, Fraser SE. The neuronal naturalist: watching neurons in their native habitat. Nat Neurosci 2001; 4:1215 - 1220
- Montagnac G, Chavrier P. Abscission accomplished by PtdIns(3)P. Nat Cell Biol 2010; 12:308 - 310
- Piel M, Nordberg J, Euteneuer U, Bornens M. Centrosome-dependent exit of cytokinesis in animal cells. Science 2001; 291:1550 - 1553
- Sagona AP, Nezis IP, Pedersen NM, Liestøl K, Poulton J, Rusten TE, et al. PtdIns(3)P controls cytokinesis through KIF13A-mediated recruitment of FYVE-CENT to the midbody. Nat Cell Biol 2010; 12:362 - 371
- Barr FA, Gruneberg U. Cytokinesis: placing and making the final cut. Cell 2007; 131:847 - 860
- Steigemann P, Gerlich DW. Cytokinetic abscission: cellular dynamics at the midbody. Trends Cell Biol 2009; 19:606 - 616
- Distel M, Babaryka A, Koster RW. Multicolor in vivo time-lapse imaging at cellular resolution by stereomicroscopy. Dev Dyn 2006; 235:1100 - 1106
- Köster RW, Fraser SE. Westerfield M, Zon L, Detrich H. Time-lapse microscopy of brain development. Essential Zebrafish Methods 2009; Philadelphia Elsevier Press 293 - 323
- Distel M, Wullimann MF, Koster RW. Optimized Gal4 genetics for permanent gene expression mapping in zebrafish. Proc Natl Acad Sci USA 2009; 106:13365 - 13370
- Scott EK, Mason L, Arrenberg AB, Ziv L, Gosse NJ, Xiao T, et al. Targeting neural circuitry in zebrafish using GAL4 enhancer trapping. Nat Methods 2007; 4:323 - 326
- Asakawa K, Kawakami K. Targeted gene expression by the Gal4-UAS system in zebrafish. Dev Growth Differ 2008; 50:391 - 399
- Distel M, Hocking JC, Volkmann K, Köster RW. The centrosome neither persistently leads migration nor determines the site of axonogenesis in migrating neurons in vivo. J Cell Biol 2010; 191:875 - 890
- Alexandre P, Reugels AM, Barker D, Blanc E, Clarke JD. Neurons derive from the more apical daughter in asymmetric divisions in the zebrafish neural tube. Nat Neurosci 2010; 13:673 - 679
- Reugels AM, Boggetti B, Scheer N, Campos-Ortega JA. Asymmetric localization of Numb: EGFP in dividing neuroepithelial cells during neurulation in Danio rerio. Dev Dyn 2006; 235:934 - 948
- von Trotha JW, Campos-Ortega JA, Reugels AM. Apical localization of ASIP/PAR-3: EGFP in zebrafish neuroepithelial cells involves the oligomerization domain CR1, the PDZ domains and the C-terminal portion of the protein. Dev Dyn 2006; 235:967 - 977
- Geldmacher-Voss B, Reugels AM, Pauls S, Campos-Ortega JA. A 90-degree rotation of the mitotic spindle changes the orientation of mitoses of zebrafish neuroepithelial cells. Development 2003; 130:3767 - 3780
- Dubreuil V, Marzesco AM, Corbeil D, Huttner WB, Wilsch-Bräuninger M. Midbody and primary cilium of neural progenitors release extracellular membrane particles enriched in the stem cell marker prominin-1. J Cell Biol 2007; 176:483 - 495
- Steigemann P, Wurzenberger C, Schmitz MH, Held M, Guizetti J, Maar S, et al. Aurora B-mediated abscission checkpoint protects against tetraploidization. Cell 2009; 136:473 - 484
- Jonsdottir AB, Dirks RW, Vrolijk J, Ogmundsdottir HM, Tanke HJ, Eyfjörd JE, et al. Centriole movements in mammalian epithelial cells during cytokinesis. BMC Cell Biol 2010; 11:34