Abstract
The Archaea constitute the third domain of life, a separate evolutionary lineage together with the Bacteria and the Eukarya.1 Species belonging to the archaea contain a surprising mix of bacterial (metabolism, lifestyle, genomic organisation) and eukaryotic (replication, transcription, translation) features.2 The archaeal kingdom comprises two main phyla, the Crenarchaeota and the Euryarchaeota. Regarding the cell division process in archaeal species (reviewed in ref. 3), members of the Euryarchaeota rely on an FtsZ-based cell division mechanism4 whereas, previously, no division genes had been detected in the crenarchaea. However, we recently reported the discovery of the elusive cell division machinery in crenarchaea from the genus Sulfolobus.5 The minimal machinery consists of three genes, which we designated cdvA, B and C (for cell division), organized into an operon that is widely conserved among crenarchaea. The gene products polymerize between segregating nucleoids at the early mitotic stage, forming a complex that remains associated with the leading edge of constriction throughout cytokinesis. Interestingly, CdvB and CdvC were shown to be related to the eukaryotic ESCRT-III protein sorting machinery (for a review on the ESCRT complex, see ref. 6), indicating shared common ancestry and mechanistic similarities to endosomal vesicle formation and viral (HIV) budding in eukaryotes. We also demonstrated that the cdv operon is subject to checkpoint-like regulation, and that the genes display a complementary phylogenetic distribution within the Archaea domain relative to FtsZ-dependent division systems5. Here, the findings are further explored and discussed, and topics for further investigation are suggested.
Multiple CdvB Paralogs in Crenarchaeal Genomes
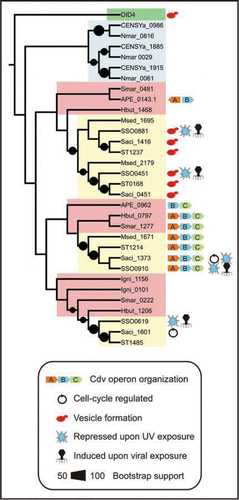
All crenarchaea that harbor the new Cdv machinery contain multiple copies of CdvB-encoding genes, resembling the situation in eukaryotes that contain an even higher number of paralogs. The respective roles for each of these CdvB paralogs are currently unclear, and present an interesting challenge for further studies. The evolutionary trajectory that gave rise to the current distribution of CdvB homologs in crenarchaeal species appears to be complex, indicating several (ancient) duplication events and perhaps also cases of horizontal gene transfer (). Irrespective of duplications and transfers, it is plausible that the Cdv-based system was present in the last common ancestor of the Crenarchaeota. Furthermore, the two CdvB clades represented by S. acidocaldarius proteins Saci_0451 and Saci_1416 appear to share a common origin (), originating from a more recent duplication event. Interestingly, these CdvB homologs have been implicated in formation of membrane vesicles,Citation7 and might be involved in excretion of enzymes into the environment. These proteins may therefore represent functional analogs of the CHMP proteins, which play important roles in ESCRT-mediated vesicle formation in eukaryotes. Moreover, all CdvB paralogs present in Sulfolobus solfataricus are significantly induced upon infection with Sulfolobus Turreted Icosahedral Virus (STIV).Citation8 One of the paralogs, SSO0881, has been found to be consistently present in STIV virion particles,Citation9 suggesting that this protein, and perhaps the other CdvB paralogs as well, might play a role in viral budding. Another interesting result from the phylogenetic analysis is that the paralogs that belong to the Cdv operonCitation5 form a monophyletic clade (except for one of the A. pernix CdvB proteins), supporting a role in cell division for all proteins within this clade.
The Cdv genes were found to be downregulated upon exposure to UV radiation in two independent studies,Citation10,Citation11 indicating a checkpoint-like responseCitation5 that enables the cell to repair DNA damage before resumption of cell cycle progression. Interestingly, several of the other CdvB paralogs in Sulfolobus are also among the most downregulated genes upon UV exposure (), suggesting that these proteins also may play a role in the division process, further supported by the observation that some of the Cdv paralogs may interact with each other.Citation12 In addition, the fact that both the UV-repression and STIV-induction (above) appear to affect the same gene set () provides a striking parallel that may indicate common regulatory features.
The three CdvB copies encoded in each of the genomes of the Nitrosopumilus maritimus and Cenarchaeum symbiosum form a monophyletic clade, indicating two consecutive duplication events within this lineage. Given that the genomes of these low-temperature crenarchaea also encode an FtsZ homolog, it remains to be shown whether the Cdv machinery is employed in the process of cell division in these organisms, or whether it may play a role e.g., in vesicle formation (see below).
Altogether, the different CdvB homologs encoded by crenarchaeal genomes appear to be occupied with a wide variety of cellular functions, ranging from cell division to virion release and vesicle formation. Additional experimental work will be necessary to decipher the precise roles of CdvB proteins in each of these processes.
Lipid Requirements in Vesicle Formation
Lysobisphosphatidic acid has been suggested to induce the formation of multivesicular liposomes in eukaryotes.Citation13 However, since this compound appears to be absent in yeast,Citation14 the lipid requirements for vesicle formation are under discussion. Archaeal cell membranes contain a unique combination of the glycerol-1-phosphate stereoisomer ether-linked to various phytanyl lipid derivatives, sometimes even crosslinked into tetraether molecules within a partly monolayer membrane. Archaeal ESCRT-III homologs are, thus, able to function within an entirely different lipid environment as compared to endosomal vesicle formation and virus budding in eukaryotic membranes, which contain fatty acids estrified to glycerol-3-phosphate. This indicates considerable flexibility in the lipid requirements for vesicle formation, at least regarding ESCRT-III mediated processes.
Archaea as Model Systems for Endosomal Sorting and Viral Budding
The relative simplicity of archaeal species compared to multicellular organisms with many-fold larger gene sets underscores their potential usefulness as model systems for certain aspects of eukaryotic cell biology. Thus, insights into core mechanistic features of ESCRT-III mediated vesicle formation may be gained from an in vitro reconstituted Sulfolobus system dependent upon significantly fewer components, as compared to the eukaryotic corresponding process. Moreover, archaea that utilize the Cdv machinery may also provide tractable models for central aspects of the budding process of HIV,Citation15 HBVCitation16 and other viruses that recruit ESCRT-III functions during release from the host cell. This aspect would become even more relevant if a role for the Cdv proteins, or any of the additional CdvB paralogs, could be demonstrated also in archaeal virus biology, and the induction of the Sulfolobus solfataricus cdv operon, as well as of the other CdvB paralogs, during infection with STIV,Citation8 indicates that this may be the case. Thus, further investigation into the budding of STIV and other archaeal viruses could potentially result in mechanistic and regulatory insights of relevance also for eukaryotic virus biology.
Cell Division in Thermoproteales
An interesting question for future investigation concerns the nature of the unidentified cell division machinery within the Thermoproteales order. Comparative genomics and other bioinformatics approaches may provide initial clues, but experimental evidence will ultimately be needed. The organisms within this order are, however, even less amenable to experimental approaches than Sulfolobus species, being strictly anaerobic (except for certain Pyrobaculum species) in addition to being hyperthermophiles, often with even higher growth temperature optima than Sulfolobus, and few research groups are able to cultivate and perform well-controlled physiological studies on such species. Furthermore, no efficient genetic systems are available for these organisms. Still, a systematic search for yet another way to carry out a process as central as cell division is likely to yield novel unexpected physiological and evolutionary insights.
Cell Division in Low-Temperature Crenarchaea
The fact that the cdv and ftsZ cell division genes are simultaneously present in two low-temperature crenarchaeal species, Nitrosopumilus maritimus and Cenarchaeum symbiosum, is intriguing. Do these species combine features of Cdv- and FtsZ-dependent cell division mechanisms into a single process?Citation17 If so, this opens up a new interesting field for investigation. Alternatively, do new surprises await discovery in these organisms, perhaps in the shape of internal membrane systems, maybe even related to eukaryotic ER, Golgi or lysosomal systems, or concerning processes mediating export of lipid vesicles? Studies of archaea have resulted in a wide range of unexpected findings, often in terms of characteristics related to eukaryotic features, and additional surprises may well await discovery through investigations into the issues outlined above.
Figures and Tables
Figure 1 Phylogenetic tree of CdvB paralogs encoded by crenarchaeal genomes. The tree was inferred from the CdvB alignment in reference 5, using RAxML and the WAGPROTGAMMA model for protein evolution.Citation18 The yeast DID4 protein, which is part of the ESCRT-III machinery, was used to root the tree. Bootstrap support values above 50 are plotted as black circles. The tree is annotated with the following biological features: Cell cycle-specific expression;Citation19 Induction upon infection by Sulfolobus turreted icosahedral virus;Citation8 Repression upon UV exposureCitation10,Citation11 and presence in secreted membrane vesicles.Citation7 Proteins are annotated using their locus tags, which includes the following species abbreviations: APE, Aeropyrum pernix; CENSYa, Cenarchaeum symbiosum; Hbut: Hyperthermus butylicus; Igni: Ignicoccus hospitalis; Msed: Metallosphaera sedula; Nmar: Nitrosopumilus maritimus; Sac: Sulfolobus acidocaldarius; Smar: Staphylothermus marinus F1; SSO: Sulfolobus solfataricus P2; ST: Sulfolobus tokodaii. Species belonging to the Sulfolobales, Desulfurococcales and Cenarchaeales orders are shaded yellow, red and blue, respectively. The only eukaryotic sequence (yeast protein DID4) is shaded green.
Acknowledgements
We are grateful to Jim Hurley for bringing attention to the role of lipids in ESCRT-III mediated vesicle formation. This work was supported by the Swedish Research Council (Rolf Bernander) and by a Marie Curie Intra-European Fellowship (Thijs J.G. Ettema).
Addendum to:
References
- Woese CR, Kandler O, Wheelis ML. Towards a natural system of organisms: proposal for the domains Archaea, Bacteria and Eucarya. Proc Natl Acad Sci USA 1990; 87:4576 - 4579
- Makarova KS, Aravind L, Galperin MY, Grishin NV, Tatusov RL, Wolf YI, Koonin EV. Comparative genomics of the Archaea (Euryarchaeota): evolution of conserved protein families, the stable core, and the variable shell. Genome Res 1999; 9:608 - 628
- Lundgren M, Bernander R. Archaeal cell cycle progress. Curr Opin Microbiol 2005; 8:662 - 668
- Wang X, Lutkenhaus J. FtsZ ring: the eubacterial division apparatus conserved in archaebacteria. Mol Microbiol 1996; 21:313 - 319
- Lindås AC, Karlsson EA, Lindgren MT, Ettema TJG, Bernander R. A unique cell division machinery in the Archaea. Proc Natl Acad Sci USA 2008; 105:18942 - 18946
- Hurley JH. ESCRT complexes and the biogenesis of multivesicular bodies. Curr Opin Cell Biol 2008; 20:4 - 11
- Ellen AF, Albers SV, Huibers W, Pitcher A, Hobel CF, Schwarz H, et al. Proteomic analysis of secreted membrane vesicles of archaeal Sulfolobus species reveals the presence of endosome sorting complex components. Extremophiles 2009; 13:67 - 79
- Ortmann AC, Brumfield SK, Walther J, McInnerney K, Brouns SJ, van de Werken HJ, et al. Transcriptome analysis of infection of the archaeon Sulfolobus solfataricus with Sulfolobus turreted icosahedral virus. J Virol 2008; 82:4874 - 4883
- Maaty WS, Ortmann AC, Dlakic M, Schulstad K, Hilmer JK, Liepold L, et al. Characterization of the archaeal thermophile Sulfolobus turreted icosahedral virus validates an evolutionary link among double-stranded DNA viruses from all domains of life. J Virol 2006; 80:7625 - 7635
- Fröls S, Gordon PM, Panlilio MA, Duggin IG, Bell SD, Sensen CW, et al. Response of the hyperthermophilic archaeon Sulfolobus solfataricus to UV damage. J Bacteriol 2007; 189:8708 - 8718
- Götz D, Paytubi S, Munro S, Lundgren M, Bernander R, White MF. Responses of hyperthermophilic crenarchaea to UV irradiation. Genome Biol 2007; 8:220
- Samson RY, Obita T, Freund SM, Williams RL, Bell SD. A role for the ESCRT System in cell division in Archaea. Science 2008; 322:1710 - 1713
- Matsuo H, Chevallier J, Mayran N, Le Blanc I, Ferguson C, Faure J, et al. Role of LBPA and Alix in multivesicular liposome formation and endosome organization. Science 2004; 303:531 - 534
- Hurley JH, Emr SD. The ESCRT complexes: structure and mechanism of a membrane-trafficking network. Annu Rev Biophys Biomol Struct 2006; 35:277 - 298
- Morita E, Sundquist WI. Retrovirus budding. Annu Rev Cell Dev Biol 2004; 20:395 - 425
- Lambert C, Döring T, Prange R. Hepatitis B virus maturation is sensitive to functional inhibition of ESCRT-III, Vps4, and g2-adaptin. J Virol 2007; 81:9050 - 9060
- Cann IK. Cell sorting protein homologs reveal an unusual diversity in archaeal cell division. Proc Natl Acad Sci USA 2008; 105:18653 - 18654
- Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 2006; 22:2688 - 2690
- Lundgren M, Bernander R. Genome-wide transcription map of an archaeal cell cycle. Proc Natl Acad Sci USA 2007; 104:2939 - 2944