Abstract
Altered levels of global DNA methylation and gene silencing through methylation of promoter regions can impact cancer risk, but little is known about their environmental determinants. We examined the association between lifestyle factors and levels of global genomic methylation and IL-6 promoter methylation in white blood cell DNA of 165 cancer-free subjects, 18–78 years old, enrolled in the COMIR (Commuting Mode and Inflammatory Response) study, New York, 2009–2010. Besides self-administrated questionnaires on diet and physical activity, we measured weight and height, white blood cell (WBC) counts, plasma levels of high sensitivity C-reactive protein (hs-CRP), and genomic (LINE-1) and gene-specific methylation (IL-6) by pyrosequencing in peripheral blood WBC. Mean levels of LINE-1 and IL-6 promoter methylation were 78.2% and 57.1%, respectively. In multivariate linear regression models adjusting for age, gender, race/ethnicity, body mass index, diet, physical activity, WBC counts and CRP, only dietary folate intake from fortified foods was positively associated with LINE-1 methylation. Levels of IL-6 promoter methylation were not significantly correlated with age, gender, race/ethnicity, body mass index, physical activity or diet, including overall dietary patterns and individual food groups and nutrients. There were no apparent associations between levels of methylation and inflammation markers such as WBC counts and hs-CRP. Overall, among several lifestyle factors examined in association with DNA methylation, only dietary folate intake from fortification was associated with LINE-1 methylation. The long-term consequence of folate fortification on DNA methylation needs to be further evaluated in longitudinal settings.
Keywords: :
Introduction
DNA methylation is an epigenetic event that may lead to cancer and other human diseases by altering gene expression. Global DNA hypomethylation and gene-specific hypermethylation are the two types of DNA methylation changes often implicated in carcinogenesis.Citation1 Global DNA hypomethylation often occurs in repetitive elements of the genome such as long interspersed repeat sequences (LINE-1) and is associated with genomic instability and chromosome abnormalities.Citation2,Citation3 Gene-specific methylation occurs at specific regions of the gene such as gene promoters and can either silence or activate the gene.Citation4 An increasing number of studies have reported a reduced level of global DNA methylation in the peripheral blood of cancer patients compared with that of healthy controls.Citation5-Citation10 Studies also found cancer-related genes are hypermethylated or hypomethylated in the white blood cells of cancer patients.Citation11-Citation16 These data suggest that global DNA hypomethylation and gene-specific methylation may serve as epigenetic markers for cancer risk.
As aberrant DNA methylation is associated with cancer and other human diseases, there are growing interests in determining how environmental exposures may affect patterns of DNA methylation. Previous studies found older monozygotic twins were more diverse in DNA methylation patterns than were younger monozygotic twins.Citation17 There is also evidence that DNA methylation patterns in individuals change over time,Citation18 supporting aging and/or external environment factors affecting DNA methylation. However, which environmental exposures, among many that individuals are exposed to over the life course, affect patterns of DNA methylation remains elusive.
Chronic inflammation constitutes an important mechanism for cancer.Citation19 The specific pathways by which inflammation causes cancer are not fully understood. Interleukin (IL-6) is a pro-inflammatory cytokine released by inflammatory cells and can activate intracellular transcription factors leading to tumorigenesis. There are some suggestions that IL-6 may stimulate tumor proliferation by altering patterns of DNA methylation,Citation20-Citation22 and IL-6 production itself is regulated by methylation of the IL-6 gene promoter.Citation23 The association between inflammation and DNA methylation has not been well explored. In this study, we examined how lifestyles (diet and physical activity) were associated with global DNA methylation using LINE-1 methylation as a surrogate and promoter methylation of IL-6 in peripheral blood, as well as the association between inflammation markers and methylation.
Results
LINE-1 methylation and promoter methylation of IL-6 in white blood cells (WBC) were approximately normally distributed. Median and mean levels of LINE-1 methylation were 78.3% and 78.2%, respectively (range: 72.5–85%); for IL-6 promoter methylation, the median and mean levels were 55.9% and 57.1%, respectively (range: 33.4–96%).
Age, gender and race/ethnicity were not significantly associated with WBC LINE-1 methylation (). However, overweight or obese subjects (BMI ≥ 25 kg/m2) had a significantly lower level of WBC LINE-1 methylation than those with healthy weight (BMI < 25 kg/m2) (77.8 vs. 78.5%, p = 0.03). Mode of commute (car drivers vs. public transportation users), dietary patterns (prudent and Western diets), adherence to USDA dietary guidelines, adherence to physical activity guidelines (2005 USDA Dietary Guidelines for Americans for physical activity or Meet Healthy People 2010 Guidelines) and alcohol drinking were not significantly associated with LINE-1 methylation. For promoter methylation of IL-6, among multiple study characteristics examined, only prudent dietary pattern was significantly associated with IL-6 promoter methylation in white blood cells. Subjects with a higher dietary score for the prudent diet had a lower level of IL-6 promoter methylation than those with a lower prudent diet score (57.1 and 62.3% for Q1 and Q2, and 54.5 and 55.8% for Q3 and Q4, p = 0.03).
Table 1. White blood cell LINE-1 methylation (%) and IL-6 promoter methylation (%) by study characteristics; New York, 2009–2010
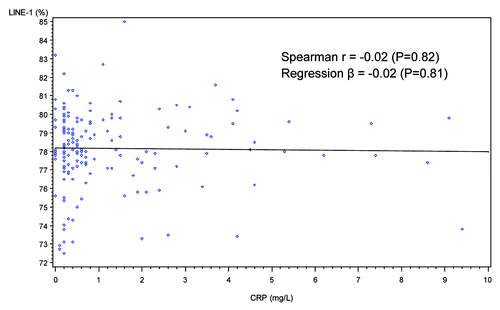
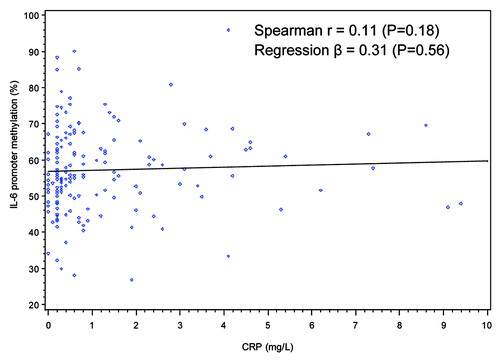
When dietary intake of individual food groups and nutrients were examined, no significant correlations were found for levels of LINE-1 methylation or IL-6 promoter methylation except that dietary intake of folate from fortified foods (such as breakfast cereals) was positively correlated with LINE-1 methylation (Spearman r = 0.21, p = 0.007) (). No significant correlations were found between DNA methylation patterns and levels of various physical activities including minutes per day of moderate leisure-time physical activities, vigorous leisure-time physical activities, job-related light activities and sedentary activities (data not shown). Two inflammation markers, WBC counts and plasma levels of hs-CRP, were not significantly correlated with LINE-1 methylation or IL-6 promoter methylation (, , and ).
Table 2. Spearman correlation coefficients between daily consumption of foods/nutrients and levels of white blood cell LINE-1 methylation and IL-6 promoter methylation, New York, 2009–2010
Figure 2. Scatter plot and simple regression line of WBC count (cells/mL) and LINE-1 methylation (%).
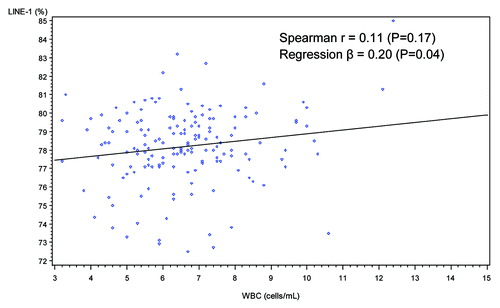
Figure 4. Scatter plot and simple regression line of WBC count (cells/mL) and IL-6 promoter methylation (%).
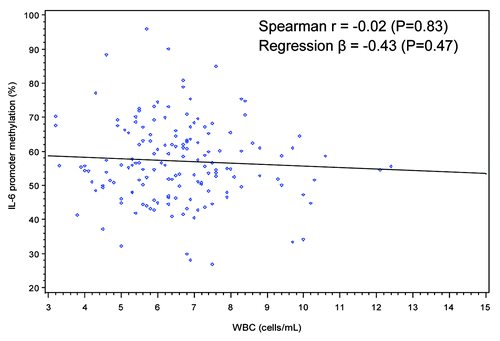
When study characteristics were examined in multivariate linear regression models, BMI was no longer significantly associated with LINE-1 methylation but a higher dietary intake of folate from fortified foods was still positively associated with LINE-1 methylation (%) (Q3 vs. Q1, β = 1.60, 95%CI: 0.54, 2.67; Q4 vs. Q1, β = 1.05, 95% CI: 0.002, 2.01, P trend = 0.03). For IL-6 promoter methylation, there was no significant trend found. Consistent with findings from the univariate analysis, age, gender, race/ethnicity, adherence to physical activity guidelines, alcohol drinking, WBC counts and plasma levels of hs-CRP were not associated with LINE-1 or IL-6 methylation.
Discussion
In a study of 165 cancer-free adults aged 18–78 y, we examined levels of WBC global methylation and promoter methylation of IL-6 in association with demographic and behavioral risk factors such as diet and physical activity as well as inflammation markers. We found a positive association between dietary folate intake from fortified foods and WBC LINE-1 methylation.
Demographic variables such as age, gender and race/ethnicity have been proposed to affect global DNA methylation. The overall impact of aging on methylation has been suggested to reduce the global level of genomic DNA methylation.Citation24 Age, as a surrogate measure for the aging process or environmental exposure over the life course, has been examined in association with WBC LINE-1 methylation in several studies. Most studies, however, do not support an age-dependent effect of WBC LINE-1 methylation.Citation25 In this young population (mean age = 29.4 y), we did not find a significant association between age and WBC LINE-1 methylation. Because the study population is relatively young, the lack of an age-dependent effect of LINE-1 methylation could be due to the overall young age. Others suggested that age may only account for a small proportion of the inter-individual variation of global methylation in humans.Citation25
Several studies documented a significantly lower level of LINE-1 methylation in women than in men.Citation25 Some suggested that women may have a higher folate requirement than men due to regular loss of red blood cells through menstruation.Citation7 X chromosome inactivation in women may also have depleted resources required for properly methylating autosomal loci.Citation26 Or the difference could be due to the copy number variation in LINE-1 in X and Y chromosomes.Citation26 In this study population, however, levels of LINE-1 methylation did not differ by gender. Further analyses revealed that women reported a significantly higher level of dietary folate intake than did men (mean daily folate intake: 44.2 vs. 35.4 µg/kJ/day, p = 0.005). The higher folate intake in women may have compensated the lower LINE-1 methylation associated with the female gender. We previously found a lower level of WBC LINE-1 methylation in non-Hispanic blacks and Hispanics as compared with non-Hispanics in cancer-free adults aged 45–75 y.Citation27 However, findings on whether LINE-1 methylation differs by race/ethnicity have been inconsistent.Citation25 We did not find a difference in LINE-1 methylation by race/ethnicity in this study. The impact of race/ethnicity on WBC global methylation warrants further investigation.
Nutrients that provide methyl-group donors such as folate, vitamin B12, vitamin B6 and methionine are important players in DNA methylation. In line with previous findings,Citation7,Citation9,Citation28 we did not find dietary folate intake or dietary intake of other vitamins in one-carbon metabolism was associated with global methylation in WBCs. However, when different food sources of dietary folate were examined, we found a significantly higher level of LINE-1 methylation associated with higher dietary folate intake from fortified foods (e.g., breakfast cereal) but not with folate intake from natural foods (e.g., leafy green vegetables). This finding is not surprising given that the National Health and Nutrition Examination Surveys (NHANES) found that blood folate levels have been substantially increased in the US population after folate fortification was implemented.Citation29 The synthetic form of folate used in fortification is more bioactive than the natural form in providing one-carbon units for DNA methylation. Folate fortification may have significantly increased the levels of global methylation in the general population. Studies investigating the association between obesity and global WBC DNA methylation are limited. Most studies reported no associations between BMI and global DNA methylation.Citation30,Citation31 Although we found a lower level of LINE-1 methylation in overweight/obese subjects, the association became statistically insignificant in multivariate regression models. We previously reported a positive trend of higher levels of LINE-1 methylation with higher levels of physical activity in a middle-aged cancer-free population, and the association became statistically insignificant after multivariate adjustment.Citation30 In this study, neither different types of physical activity (e.g., leisure-time, household physical activity and job-related physical activities) nor adherence to physical activity guidelines was associated with LINE-1 methylation.
In addition to global WBC DNA methylation, gene-specific methylation of cancer-related genes such as tumor-suppressor genes has also been proposed as an epigenetic marker for cancer risk. Previous studies showed significant associations between gene-specific methylation and risks of breast and colon cancer.Citation11-Citation16 IL-6 is a prominent pro-inflammatory marker and the key inducer for CRP production. Promoter methylation of IL-6 affects gene regulation and has been implicated in the pathogenesis of rheumatoid arthritis and other autoimmune diseases.Citation23 A previous study showed that IL-6 induced significant global LINE-1 hypomethylation as well as CpG promoter methylation of several tumor suppressor genes in oral cancer cells,Citation21 suggesting inflammation can potentially affect cancer risk by altering patterns of DNA methylation. However, the two inflammation markers measured in this study (i.e., WBC count and hs-CRP) were not associated with IL-6 promoter or LINE-1 methylation. Further studies are required to disentangle how methylation status may affect or be affected by inflammation. Our study detected a positive association between the second quartile of the prudent dietary pattern (Q2) and levels of IL-6 promoter methylation when comparing to the lowest quartile (Q1). A higher quartile of the prudent dietary pattern corresponds to a higher adherence to this pattern. However, the positive association was confined to the second quartile. The lack of associations for the third and fourth quartiles does not support a consistent effect of the prudent dietary pattern on IL-6 promoter methylation. The positive finding may only be due to chance.
Our study has limitations. We measured LINE-1 methylation as a surrogate for global DNA methylation by pyrosequencing; therefore, our results are not directly comparable to those assessing other repetitive elements such as ALU or directly quantifying global methyl-cytosine content. However, previous studies showed reasonable correlations between methylation at LINE-1 and ALU elements and total methyl-cytosine content,Citation32 and pyrosequencing has been used extensively to measure global DNA methylation with a good reliability.Citation32 Nevertheless, it has not been established how adequately this surrogate marker reflects true genome-wide methylation levels. For the assessment of IL-6 promoter methylation, the variability across the sites targeted within the promoter, as indicated by the coefficient of variation, may have reduced the robustness of the designed assay to capture the small differences to be expected within this setting. Although the difference in LINE-1 methylation associated with dietary folate intake from fortified foods detected in this study was small (~1%), a small difference in LINE-1 methylation has been significantly associated with cancer and environmental exposures in previous studies.Citation7,Citation28,Citation33 We measured methylation in total WBC. Diseases, including cancer, may produce shifts in leukocyte populations, such as neutrophilia or lymphopenia.Citation34 Each lineage of white blood cells is methylated on different regions,Citation35,Citation36 and lineage specific differences may have been blurred if some cells become hypomethylated and others become hypermethylated. However, such methylation heterogeneity may be more likely to occur in the presence of leukocytosis, which is rare in this young and healthy population. Infections, drugs and exercise are among the factors that can also impact WBC populations.Citation25 While our subjects were generally healthy, unmeasured changes in specific WBC population could have affected our results. IL-6 is produced by T-cells and macrophages; increases in these cell types could lead to decreased WBC methylation levels. We assessed at least 10 dietary and lifestyle risk factors in association with two methylation outcomes, and our results may be subject to false positive findings due to multiple comparisons. Studies with a larger sample size are needed to confirm our findings. Finally, this study is cross-sectional. Further studies with a longitudinal design such as randomized intervention trails are required to suggest a causal relationship.
Overall, among several lifestyle factors examined in association with DNA methylation, dietary folate intake from fortification was the only one that showed a significant association with LINE-1 methylation. This may have important implications given that previous randomized controlled trials (RCTs) found an increased risk of colorectal and prostate cancers associated with folic acid supplementation in patients with adenomas.Citation37,Citation38 A higher incidence of all cancers and all-cause mortality were also reported in patients with ischemic heart diseases treated with folic acid plus vitamin B12.Citation39 Although fortification provides a lower dose of folic acid compared with that of supplementation (e.g., 400 μg folic acid per ¾ cup of breakfast cereals through fortification vs. 800–1,000 μg daily supplementation of folic acid in RCTs), fortification is incorporated into an individual’s daily diet with decades of exposure. Our findings suggest that folate fortification can potentially increase levels of global DNA methylation in a cancer-free population. The long-term consequence of prolonged exposure to additional folate through fortification needs to be further evaluated. Studies in prospective settings are needed to investigate the impact of the folate fortification on both global DNA methylation and promoter methylation of tumor-suppressor genes.
Patients and Methods
Study population
The Commuting Mode and Inflammatory Response (COMIR) study was conducted in 2009–2010 to assess how lifestyle factors, inflammatory and epigenetic markers differ by the mode of commuting.Citation40 A total of 180 people commuting to a college campus in Queens, NY, either by car or by public transportation, for five days/week were recruited. Commuters were identified from a campus-wide survey about commuting habits. Those who had recently used anti-inflammatory drugs, had work-related exposure to air pollutants or were current smokers were excluded.
Study participants aged 18–78 y completed a 108-item Block food frequency questionnaire (FFQ) for habitual dietary intake and a Block adult energy expenditure survey (EES) for physical activity. The Block FFQ asks study participants about their usual dietary intake of 110 food items during the last year. For each food item, the Block FFQ uses eight categories to assess frequency (never or hardly ever, once per month, 2–3 times/month, once per week, 2–3 times/week, 4–6 times/week, once per day, and ≥ 2 times/day) and three categories to assess portion size. The Block FFQ was previously validated with three 24-h dietary recalls and generally had high correlations for most nutrients.Citation41 The Block EES for physical activity assesses the frequency and duration of 26 leisure-time physical activities, job-related and home activities during the past year. Included physical activities are derived from the most relevant daily-life and leisure time activities determined by the National Human Activities Patterns Survey.Citation42 The Block EES for physical activities uses six categories (rarely or never, a few times a month, 1–2 times/week, 3–4 times/week, 5–6 times/week and everyday) to assess frequency and seven categories (> 30 min, 30–59 min, 1 h, 1.5 h, 2 h, 3–4 h and > 4 h) to assess duration. The validation study showed that the percent of body fat predicted by levels of physical activity estimated from this questionnaire correlated well with the percent body fat observed.Citation43 The FFQs and EESs completed by study participants were sent to NutritionQuest® for data processing after initial quality check at Queens College of CUNY.
Weight and height were measured using a Detecto® medical scale and gauge by trained research staff. A 15 ml non-fasting blood sample was collected from each participant by research nurses using venipuncture; 8 ml was sent immediately to a commercial clinical lab (Quest) for WBC count (cells/mm3) measurement. The remaining blood sample collected into an EDTA tube was shipped in a refrigerated box to the laboratory at Columbia University. Upon arrival, plasma and WBCs were isolated and stored at -80°C. One vial of plasma was sent to Quest for hs-CRP (mg/dl) measurement. The isolated WBCs were used to measure DNA methylation at Columbia University.
Methylation analysis
DNA was extracted from the WBCs using FlexiGene DNA Kits (Qiagen). Bisulfite modification was conducted using an EZ DNA Methylation-Gold kit (Zymo Research) following the manufacturer’s recommendations. For LINE-1, the PCR primers and sequencing probe used were previously described.Citation33 For IL-6, the PCR primers and sequencing probe were designed to target sites within a CpG island located in the promoter region of the gene using the Pyromark Assay Design Software Version 2.0 (Qiagen). The sequences were as follows: TTTTGAGAAAGGAGGTGGGTAG (Forward PCR primer), ACCCCCTTAACCTCAAATCTACAATACTCT (5′ biotinylated Reverse PCR primer), and AAGGAGGTGGGTAGG (Sequencing primer). The biotinylated PCR products were purified and pyrosequencing was run on a PyroMark Q24 (Qiagen). We used non-CpG cytosine residues as internal controls to verify efficient sodium bisulfite DNA conversion, and universal unmethylated (whole genome amplified) and methylated DNA (CpGenome Universal Methylated DNA, Millipore) were run as controls. Methylation quantification was performed using the PyroMark Q24 1.010 software. The degree of methylation was expressed for each DNA locus as percentage methylated cytosine over the sum of methylated and unmethylated cytosine. For LINE-1 values across the 3 CpG sites were averaged while for IL-6 values for the 6 sites were averaged.
Statistical analysis
Among the 180 subjects enrolled in the COMIR study, 15 did not have DNA methylation results due to low DNA yields or low quality calls. This left 165 subjects for the final analysis. We first examine mean levels of LINE-1 methylation and IL-6 promoter methylation by study characteristics including age, sex, race/ethnicity, body mass index, mode of commute, diet, physical activity and alcohol drinking, using analysis of variance (ANOVA) (). For diet, we used factor analysis (principal factor method) to derive dietary patterns from the frequency consumption of 13 food groups estimated from the Block FFQ.Citation44 Two dietary patterns were determined using the scree test, the proportion of variance accounted, and the interpretability criteria: a prudent diet characterized by a high intake of vegetables and fruits, and a western diet characterized by a high intake of meats, total grains and dairy. Scoring coefficients were computed by multiplying the matrix of factor loadings by the matrix of eigenvalues. Subjects’ individual factor scores were then estimated for the two dietary patterns by summing the frequency consumption of each food group weighted by their scoring coefficients. Therefore, factor scores for a particular dietary pattern rank individuals on the basis of how well they follow that dietary pattern. For each dietary pattern, participants were categorized into quartiles of ranked factor scores, with higher quartiles corresponding to higher factor scores. In addition, we evaluated the adherence to the US. Department of Agriculture (USDA) 2005 Dietary Guidelines for Americans (2005 DGA) using the Dietary Guideline Adherence Index (DGAI) adapted from the method developed by Jacque et al.Citation45 Previous studies showed significant associations between DGAI and insulin resistance and metabolic syndrome.Citation46,Citation47 Quartiles were created for DAGI to evaluate its association with DNA methylation. Grams per day of alcohol drinking were also derived from the Block FFQ and alcohol drinking was categorized based on conventional cut-points (never, 0–15, 15–30, > 30 g/day) that correspond to non-drinkers, < 1 drink/day, 1–2 drinks/day and more than 2 drinks/day.
Table 3. Differences in percentages of LINE-1 methylation (%) and IL-6 promoter methylation (%) according to study characteristics, New York, 2009–2010a
For physical activity, daily frequency of each type (occupational, leisure and home-based) and intensity (moderate and vigorous) of physical activities (minutes/day) was estimated from the reported frequency and duration in the Block EES. We examined adherence to physical activity guidelines by assessing whether participants met the 2005 DGA for physical activity (i.e., engaged in approximately 60 min of moderate- to vigorous-intensity activity on most days of the week) and whether participants met Healthy People 2010 Guidelines for physical activity (i.e., engaged in moderate physical activity for at least 30 min on at least five days a week, or engaged in vigorous physical activity for 20 min on at least three days per week).
We then evaluated the correlation of WBC DNA methylation (LINE-1 and IL-6 promoter methylation) levels with dietary intake of individual food groups and nutrients, and with levels of various physical activities. Because the dietary intake of most nutrients and some food groups did not follow a normal distribution, the Spearman correlation coefficients were assessed. The correlation between levels of DNA methylation and the two inflammation markers (e.g., WBC count and hs-CRP) was also assessed using Spearman correlation coefficients. Last, we examined the association between WBC DNA methylation and study characteristics including diet, physical activity and the two inflammation markers using multivariate linear regression. Levels of CPR was categorized into two groups (< 3 vs. ≥ 3 mg/L) and this cut point was chosen based on previous findings that an elevated CRP > 3 mg/L was associated with increased risk of coronary heart diseases.Citation48 Levels of WBC was categorized into four groups (< 5,500, 5500–6500, 6500–7400, ≥ 7400/μL) based on previous findings showing circulating WBC associated with subsequent cancer mortality.Citation49 Since LINE-1 and IL-6 promoter methylation were approximately normally distributed, no transformations were applied. Among multiple dietary factors, only dietary patterns and dietary folate intake from fortified foods were included in regression models because these two dietary factors showed significant correlations with DNA methylation in the univariate analyses. All statistical analyses were performed using SAS (version 9.1; SAS Institute).
Funding
CIRG2009, NIEHS Center ES009089, UNTHSC School of Public Health/Institute for Cancer Research Seed Program, Joe and Jessie Crump Fund for Medical Education
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- Yoo CB, Jones PA. Epigenetic therapy of cancer: past, present and future. Nat Rev Drug Discov 2006; 5:37 - 50; http://dx.doi.org/10.1038/nrd1930; PMID: 16485345
- Sharma S, Kelly TK, Jones PA. Epigenetics in cancer. Carcinogenesis 2010; 31:27 - 36; http://dx.doi.org/10.1093/carcin/bgp220; PMID: 19752007
- Wilson AS, Power BE, Molloy PL. DNA hypomethylation and human diseases. Biochim Biophys Acta 2007; 1775:138 - 62; PMID: 17045745
- Esteller M. CpG island hypermethylation and tumor suppressor genes: a booming present, a brighter future. Oncogene 2002; 21:5427 - 40; http://dx.doi.org/10.1038/sj.onc.1205600; PMID: 12154405
- Choi JY, James SR, Link PA, McCann SE, Hong CC, Davis W, et al. Association between global DNA hypomethylation in leukocytes and risk of breast cancer. Carcinogenesis 2009; 30:1889 - 97; http://dx.doi.org/10.1093/carcin/bgp143; PMID: 19584139
- Hou L, Wang H, Sartori S, Gawron A, Lissowska J, Bollati V, et al. Blood leukocyte DNA hypomethylation and gastric cancer risk in a high-risk Polish population. Int J Cancer 2010; 127:1866 - 74; http://dx.doi.org/10.1002/ijc.25190; PMID: 20099281
- Hsiung DT, Marsit CJ, Houseman EA, Eddy K, Furniss CS, McClean MD, et al. Global DNA methylation level in whole blood as a biomarker in head and neck squamous cell carcinoma. Cancer Epidemiol Biomarkers Prev 2007; 16:108 - 14; http://dx.doi.org/10.1158/1055-9965.EPI-06-0636; PMID: 17220338
- Lim U, Flood A, Choi SW, Albanes D, Cross AJ, Schatzkin A, et al. Genomic methylation of leukocyte DNA in relation to colorectal adenoma among asymptomatic women. Gastroenterology 2008; 134:47 - 55; http://dx.doi.org/10.1053/j.gastro.2007.10.013; PMID: 18166347
- Moore LE, Pfeiffer RM, Poscablo C, Real FX, Kogevinas M, Silverman D, et al. Genomic DNA hypomethylation as a biomarker for bladder cancer susceptibility in the Spanish Bladder Cancer Study: a case-control study. Lancet Oncol 2008; 9:359 - 66; http://dx.doi.org/10.1016/S1470-2045(08)70038-X; PMID: 18339581
- Pufulete M, Al-Ghnaniem R, Leather AJ, Appleby P, Gout S, Terry C, et al. Folate status, genomic DNA hypomethylation, and risk of colorectal adenoma and cancer: a case control study. Gastroenterology 2003; 124:1240 - 8; http://dx.doi.org/10.1016/S0016-5085(03)00279-8; PMID: 12730865
- Flanagan JM, Munoz-Alegre M, Henderson S, Tang T, Sun P, Johnson N, et al. Gene-body hypermethylation of ATM in peripheral blood DNA of bilateral breast cancer patients. Hum Mol Genet 2009; 18:1332 - 42; http://dx.doi.org/10.1093/hmg/ddp033; PMID: 19153073
- Iwamoto T, Yamamoto N, Taguchi T, Tamaki Y, Noguchi S. BRCA1 promoter methylation in peripheral blood cells is associated with increased risk of breast cancer with BRCA1 promoter methylation. Breast Cancer Res Treat 2011; 129:69 - 77; http://dx.doi.org/10.1007/s10549-010-1188-1; PMID: 20882403
- Kaaks R, Stattin P, Villar S, Poetsch AR, Dossus L, Nieters A, et al. Insulin-like growth factor-II methylation status in lymphocyte DNA and colon cancer risk in the Northern Sweden Health and Disease cohort. Cancer Res 2009; 69:5400 - 5; http://dx.doi.org/10.1158/0008-5472.CAN-08-3020; PMID: 19549920
- Leaderer D, Hoffman AE, Zheng T, Fu A, Weidhaas J, Paranjape T, et al. Genetic and epigenetic association studies suggest a role of microRNA biogenesis gene exportin-5 (XPO5) in breast tumorigenesis. Int J Mol Epidemiol Genet 2011; 2:9 - 18; PMID: 21552306
- Widschwendter M, Apostolidou S, Raum E, Rothenbacher D, Fiegl H, Menon U, et al. Epigenotyping in peripheral blood cell DNA and breast cancer risk: a proof of principle study. PLoS One 2008; 3:e2656; http://dx.doi.org/10.1371/journal.pone.0002656; PMID: 18628976
- Wong EM, Southey MC, Fox SB, Brown MA, Dowty JG, Jenkins MA, et al. Constitutional methylation of the BRCA1 promoter is specifically associated with BRCA1 mutation-associated pathology in early-onset breast cancer. Cancer Prev Res (Phila) 2011; 4:23 - 33; http://dx.doi.org/10.1158/1940-6207.CAPR-10-0212; PMID: 20978112
- Fraga MF, Ballestar E, Paz MF, Ropero S, Setien F, Ballestar ML, et al. Epigenetic differences arise during the lifetime of monozygotic twins. Proc Natl Acad Sci U S A 2005; 102:10604 - 9; http://dx.doi.org/10.1073/pnas.0500398102; PMID: 16009939
- Bjornsson HT, Sigurdsson MI, Fallin MD, Irizarry RA, Aspelund T, Cui H, et al. Intra-individual change over time in DNA methylation with familial clustering. JAMA 2008; 299:2877 - 83; http://dx.doi.org/10.1001/jama.299.24.2877; PMID: 18577732
- Balkwill F, Mantovani A. Inflammation and cancer: back to Virchow?. Lancet 2001; 357:539 - 45; http://dx.doi.org/10.1016/S0140-6736(00)04046-0; PMID: 11229684
- Foran E, Garrity-Park MM, Mureau C, Newell J, Smyrk TC, Limburg PJ, et al. Upregulation of DNA methyltransferase-mediated gene silencing, anchorage-independent growth, and migration of colon cancer cells by interleukin-6. Mol Cancer Res 2010; 8:471 - 81; http://dx.doi.org/10.1158/1541-7786.MCR-09-0496; PMID: 20354000
- Gasche JA, Hoffmann J, Boland CR, Goel A. Interleukin-6 promotes tumorigenesis by altering DNA methylation in oral cancer cells. Int J Cancer 2011; 129:1053 - 63; http://dx.doi.org/10.1002/ijc.25764; PMID: 21710491
- Wehbe H, Henson R, Meng F, Mize-Berge J, Patel T. Interleukin-6 contributes to growth in cholangiocarcinoma cells by aberrant promoter methylation and gene expression. Cancer Res 2006; 66:10517 - 24; http://dx.doi.org/10.1158/0008-5472.CAN-06-2130; PMID: 17079474
- Nile CJ, Read RC, Akil M, Duff GW, Wilson AG. Methylation status of a single CpG site in the IL6 promoter is related to IL6 messenger RNA levels and rheumatoid arthritis. Arthritis Rheum 2008; 58:2686 - 93; http://dx.doi.org/10.1002/art.23758; PMID: 18759290
- Issa JP. Age-related epigenetic changes and the immune system. Clin Immunol 2003; 109:103 - 8; http://dx.doi.org/10.1016/S1521-6616(03)00203-1; PMID: 14585281
- Terry MB, Delgado-Cruzata L, Vin-Raviv N, Wu HC, Santella RM. DNA methylation in white blood cells: association with risk factors in epidemiologic studies. Epigenetics 2011; 6:828 - 37; http://dx.doi.org/10.4161/epi.6.7.16500; PMID: 21636973
- El-Maarri O, Becker T, Junen J, Manzoor SS, Diaz-Lacava A, Schwaab R, et al. Gender specific differences in levels of DNA methylation at selected loci from human total blood: a tendency toward higher methylation levels in males. Hum Genet 2007; 122:505 - 14; http://dx.doi.org/10.1007/s00439-007-0430-3; PMID: 17851693
- Zhang FF, Cardarelli R, Carroll J, Fulda KG, Kaur M, Gonzalez K, et al. Significant differences in global genomic DNA methylation by gender and race/ethnicity in peripheral blood. Epigenetics 2011; 6:623 - 9; http://dx.doi.org/10.4161/epi.6.5.15335; PMID: 21739720
- Zhang FF, Morabia A, Carroll J, Gonzalez K, Fulda K, Kaur M, et al. Dietary patterns are associated with levels of global genomic DNA methylation in a cancer-free population. J Nutr 2011; 141:1165 - 71; http://dx.doi.org/10.3945/jn.110.134536; PMID: 21525250
- McDowell MA, Lacher DA, Pfeiffer CM, Mulinare J, Picciano MF, Rader JI, et al. Blood folate levels: the latest NHANES results. NCHS Data Brief 2008:1-8.
- Zhang FF, Cardarelli R, Carroll J, Zhang S, Fulda KG, Gonzalez K, et al. Physical activity and global genomic DNA methylation in a cancer-free population. Epigenetics 2011; 6:293 - 9; http://dx.doi.org/10.4161/epi.6.3.14378; PMID: 21178401
- Zhu ZZ, Hou L, Bollati V, Tarantini L, Marinelli B, Cantone L, et al. Predictors of global methylation levels in blood DNA of healthy subjects: a combined analysis. Int J Epidemiol 2012; 41:126 - 39; http://dx.doi.org/10.1093/ije/dyq154; PMID: 20846947
- Yang AS, Estécio MR, Doshi K, Kondo Y, Tajara EH, Issa JP. A simple method for estimating global DNA methylation using bisulfite PCR of repetitive DNA elements. Nucleic Acids Res 2004; 32:e38; http://dx.doi.org/10.1093/nar/gnh032; PMID: 14973332
- Bollati V, Baccarelli A, Hou L, Bonzini M, Fustinoni S, Cavallo D, et al. Changes in DNA methylation patterns in subjects exposed to low-dose benzene. Cancer Res 2007; 67:876 - 80; http://dx.doi.org/10.1158/0008-5472.CAN-06-2995; PMID: 17283117
- Tavares-Murta BM, Mendonça MA, Duarte NL, da Silva JA, Mutão TS, Garcia CB, et al. Systemic leukocyte alterations are associated with invasive uterine cervical cancer. Int J Gynecol Cancer 2010; 20:1154 - 9; http://dx.doi.org/10.1111/IGC.0b013e3181ef8deb; PMID: 21495217
- Schmidl C, Klug M, Boeld TJ, Andreesen R, Hoffmann P, Edinger M, et al. Lineage-specific DNA methylation in T cells correlates with histone methylation and enhancer activity. Genome Res 2009; 19:1165 - 74; http://dx.doi.org/10.1101/gr.091470.109; PMID: 19494038
- Wu HC, Delgado-Cruzata L, Flom JD, Kappil M, Ferris JS, Liao Y, et al. Global methylation profiles in DNA from different blood cell types. Epigenetics 2011; 6:76 - 85; http://dx.doi.org/10.4161/epi.6.1.13391; PMID: 20890131
- Cole BF, Baron JA, Sandler RS, Haile RW, Ahnen DJ, Bresalier RS, et al, Polyp Prevention Study Group. Folic acid for the prevention of colorectal adenomas: a randomized clinical trial. JAMA 2007; 297:2351 - 9; http://dx.doi.org/10.1001/jama.297.21.2351; PMID: 17551129
- Figueiredo JC, Grau MV, Haile RW, Sandler RS, Summers RW, Bresalier RS, et al. Folic acid and risk of prostate cancer: results from a randomized clinical trial. J Natl Cancer Inst 2009; 101:432 - 5; http://dx.doi.org/10.1093/jnci/djp019; PMID: 19276452
- Ebbing M, Bønaa KH, Nygård O, Arnesen E, Ueland PM, Nordrehaug JE, et al. Cancer incidence and mortality after treatment with folic acid and vitamin B12. JAMA 2009; 302:2119 - 26; http://dx.doi.org/10.1001/jama.2009.1622; PMID: 19920236
- Morabia A, Zhang FF, Kappil MA, Flory J, Mirer FE, Santella RM, et al. Biologic and epigenetic impact of commuting to work by car or using public transportation: A case-control study. Prev Med 2012; 54:229 - 33; http://dx.doi.org/10.1016/j.ypmed.2012.01.019; PMID: 22313796
- Subar AF, Thompson FE, Kipnis V, Midthune D, Hurwitz P, McNutt S, et al. Comparative validation of the Block, Willett, and National Cancer Institute food frequency questionnaires : the Eating at America’s Table Study. Am J Epidemiol 2001; 154:1089 - 99; http://dx.doi.org/10.1093/aje/154.12.1089; PMID: 11744511
- Dong L, Block G, Mandel S. Activities Contributing to Total Energy Expenditure in the United States: Results from the NHAPS Study. Int J Behav Nutr Phys Act 2004; 1:4; http://dx.doi.org/10.1186/1479-5868-1-4; PMID: 15169563
- Block G, Jensen CD, Block TJ, Norris J, Dalvi TB, Fung EB. The work and home activities questionnaire: energy expenditure estimates and association with percent body fat. J Phys Act Health 2009; 6:Suppl 1 S61 - 9; PMID: 19998851
- Tucker KL. Dietary patterns, approaches, and multicultural perspective. Appl Physiol Nutr Metab 2010; 35:211 - 8; http://dx.doi.org/10.1139/H10-010; PMID: 20383235
- Fogli-Cawley JJ, Dwyer JT, Saltzman E, McCullough ML, Troy LM, Jacques PF. The 2005 Dietary Guidelines for Americans Adherence Index: development and application. J Nutr 2006; 136:2908 - 15; PMID: 17056821
- Fogli-Cawley JJ, Dwyer JT, Saltzman E, McCullough ML, Troy LM, Meigs JB, et al. The 2005 Dietary Guidelines for Americans and insulin resistance in the Framingham Offspring Cohort. Diabetes Care 2007; 30:817 - 22; http://dx.doi.org/10.2337/dc06-1927; PMID: 17259479
- Fogli-Cawley JJ, Dwyer JT, Saltzman E, McCullough ML, Troy LM, Meigs JB, et al. The 2005 Dietary Guidelines for Americans and risk of the metabolic syndrome. Am J Clin Nutr 2007; 86:1193 - 201; PMID: 17921402
- Shankar A, Wang JJ, Rochtchina E, Yu MC, Kefford R, Mitchell P. Association between circulating white blood cell count and cancer mortality: a population-based cohort study. Arch Intern Med 2006; 166:188 - 94; http://dx.doi.org/10.1001/archinte.166.2.188; PMID: 16432087
- Miller M, Zhan M, Havas S. High attributable risk of elevated C-reactive protein level to conventional coronary heart disease risk factors: the Third National Health and Nutrition Examination Survey. Arch Intern Med 2005; 165:2063 - 8; http://dx.doi.org/10.1001/archinte.165.18.2063; PMID: 16216995