Abstract
Germline mutations in the BRCA1 or BRCA2 genes are associated with an increased risk of breast and ovarian cancer development. Both genes are involved in DNA repair, and tumors harboring genetic defects in them are thought to be more sensitive to DNA-damaging agents used in chemotherapy. However, as only a minority of breast and ovarian cancer patients carry BRCA1 or BRCA2 mutations, few patients are likely to benefit from these pharmacogenetic biomarkers. Herein, we show that, in cancer cell lines and xenografted tumors, BRCA1 CpG island promoter hypermethylation-associated silencing also predicts enhanced sensitivity to platinum-derived drugs to the same extent as BRCA1 mutations. Most importantly, BRCA1 hypermethylation proves to be a predictor of longer time to relapse and improved overall survival in ovarian cancer patients undergoing chemotherapy with cisplatin.
Introduction
Female BRCA1 and BRCA2 mutation carriers have a significantly higher lifetime risk of breast and ovarian cancer.Citation1 BRCA1 and BRCA2 proteins play major roles in DNA double-strand-break repair through homologous recombination,Citation2 so their deficiencies can impair the capacity of cancer cells to repair DNA cross-links caused by chemotherapy drugs such as platinum-derivatives.Citation3-Citation7 Ovarian cancer accounts for more deaths than any other tumor of the female reproductive system, so there is great interest in identifying biomarkers for therapy prediction. Two independent studies reported significantly greater primary chemotherapy sensitivity to platinum-based chemotherapy agents in patients with ovarian cancer who were carriers of BRCA1 and BRCA2 germline mutations.Citation5,Citation6 In addition, tumors from carriers of BRCA1/BRCA2 germline mutations are also sensitive to poly (ADP-ribose) polymerase inhibitors (PARPis) that target the base excision repair pathway.Citation8-Citation12 However, only a minority of breast and ovarian cancer patients are BRCA1 and BRCA2 mutation carriers, so the benefit of these findings might be confined to a small subset of cases. In addition, it might be a link between BRCA1/BRCA2 defects, platinum sensitivity and response to PARPis in breast and ovarian tumors that is becoming an issue of growing interest.Citation8-Citation12 Herein, we have approached this matter from a different angle.
In the search for new potential biomarkers of sensitivity differences of human cancer to chemotherapeutic agents, the existence of aberrations in the DNA methylation patterns of cancer cells is turning out to be the most important, particularly those involving hypermethylation of the sequences called CpG islands, which are located in the promoter regions of tumor suppressor genes.Citation13 One of the most successful discoveries in this area, made by our groupCitation14 and others,Citation15 and subsequently validated worldwide,Citation16 is that hypermethylation of the DNA repair enzyme MGMT is associated with a good response to nitrosurea alkylating agents in glioma. For BRCA1, there is clear evidence that the BRCA1 gene can also undergo epigenetic inactivation in sporadic breast tumorsCitation17-Citation22 and ovarian tumorsCitation20,Citation23-Citation25 by the gain of DNA methylation in its promoter-associated CpG island. That this aberration produces a tumor with a BRCA1 phenotype was further demonstrated by showing that it gives rise to the same pattern of gene expression as seen in inherited BRCA1 mutations.Citation26 Strikingly, we and others have recently found that BRCA1 CpG island hypermethylation also predicts sensitivity to PARPis.Citation27,Citation28
We examined whether the enhanced platinum-based sensitivity observed in BRCA1/BRCA2 familial tumors is also present in sporadic BRCA1 hypermethylated tumors.
Results and Discussion
BRCA1 and BRCA2 are candidate genes for hypermethylation-associated inactivation in human cancer because a 5′-CpG island is located around the corresponding transcription start sites. To analyze the methylation status of the promoter-associated CpG islands, we screened 15 human cancer cell lines from breast (HCC-1143, MDA-MB-468, MDA-MB-468-PT, MDA-MB-468LN, MCF7, SK-BR-3, T47D, Hs578T, UACC3199, MDA-MB-231 and MDA-MB-436) and ovarian (SK-OV-3, IGR-OV1, OVCAR-3 and OVCAR-5) tumor types, using bisulfite genomic sequencing, methylation-specific PCR and pyrosequencing. BRCA2 promoter CpG island methylation was not found in any of the cases, but the breast cancer cell lines UACC3199 and HCC-38 exhibited BRCA1 CpG island promoter hypermethylation (). All normal breast tissues analyzed were completely unmethylated at the BRCA1 promoter CpG island ().
Figure 1. BRCA1 promoter CpG island hypermethylation is associated with transcriptional silencing. (A) Pyrosequencing analysis of BRCA1 CpG island demonstrates hypermethylation in UACC-3199 and HCC-38 cancer cells. (B) Bisulfite genomic sequencing of eight individual clones in the BRCA1 promoter CpG island: examples of a normal breast and the breast cancer cell line HCC-38 are shown. Presence of a methylated or unmethylated cytosine is indicated by a black or white square, respectively. Black arrows indicate the position of the bisulfite genomic sequencing primers. (C) Real-time PCR expression of the BRCA1 transcript. (D) BRCA1 expression was also determined by western blot and the β-actin protein was used as a loading control. The UACC3199 and HCC-38 breast cancer cells show a hypermethylated CpG island in association with the downregulation of the BRCA1 protein. MDA-MB-231 (wild-type) and MDA-MB-436 (mutant) are shown as positive and negative controls for BRCA1 expression.
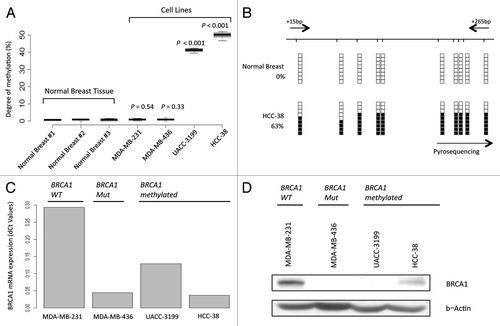
Having noted BRCA1 promoter hypermethylation in the aforementioned cancer cell lines, we assessed the association between this epigenetic aberration and the putative transcriptional inactivation of the BRCA1 gene at the RNA and protein levels. The cancer cell lines UACC3199 and HCC-38 hypermethylated at the BRCA1 CpG island had minimal expression of the BRCA1 RNA transcript, as determined by quantitative RT-PCR (), and BRCA1 protein, as determined by western blot (Anti-BRCA1 Ab-1, Calbiotech, Clone# MS110) (). The BRCA1 mutant breast cancer cell line MDA-MB-436 cell, which carries a genetic deletion, was used as a control for the lack of expression of the BRCA1 transcript and protein (). In contrast, the BRCA1 unmethylated and non-mutant MDA-MB-231 cell line expressed the BRCA1 transcript and protein ().
An increasing number of reports suggest that tumors with genetic defects in BRCA1 are more sensitive to growth inhibition and chromosomal damage upon platinum-based chemotherapy. This makes it extremely interesting to know, for clinical translational purposes, whether cancer cells with BRCA1 methylation-associated silencing also possess these functional features. First, we studied the antiproliferation effects of cisplatin and carboplatin in the four described cancer cell lines with different BRCA1 genetic/epigenetic status using the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay. BRCA1 hypermethylation (UACC3199 and HCC-38 cells) conferred the same degree of sensitivity to the two platin compounds as did the BRCA1 mutation (), while the unmethylated and non-mutated cell line (MDA-MB-231) was significantly more resistant (). Related to the formation of double-strand breaks in the DNA upon the use of the platin-derivatives, BRCA1-deficient cells (hypermethylated or mutated) experienced equally massive DNA damage, as assessed by the comet assay, when treated with cisplatin or carboplatin (). This was not observed in BRCA1-proficient cells (). It is interesting to note that BRCA1 unmethylated and non-mutated cells express increasing amounts of BRCA1 when platin is used, enabling the effective repair of induced DNA lesions, but BRCA1 hypermethylated cells are unable to experience this reactive change ().
Figure 2. BRCA1 epigenetic inactivation predicts sensitivity to platinum-based chemotherapy. (A) Cell viability assessed by the MTT assays demonstrates that methylated (UACC3199 and HCC-38) and mutant (MDA-MB-436) BRCA1 cells both exhibit enhanced sensitivity to cisplatin and carboplatin in comparison with wild type and unmethylated MDA-MB-231 breast cancer cells. The corresponding IC50 values are shown. (B) Representative comet assays show DNA damage upon cisplatin use in the BRCA1 methylated or mutated cell lines. (C) Quantification of the obtained values from the comet assay. (E) BRCA1-hypermethylated cells are not able to repair DNA damage when cisplatin is used. The values of comet assays shown in box-plots demonstrate that both methylated and mutant BRCA1 cells experience permanent DNA damage when cisplatin is used that it is not observed in BRCA1 wild type or unmethylated cells (MDA-MB-231). (D) Relative changes in tumor size of UACC3199 (BRCA1 hypermethylated) and MDA-MB-231 (BRCA1 unmethylated) cancer cells xenografted in nude mice upon cisplatin use. Values shown at 28 d after the start of the chemotherapy treatment.
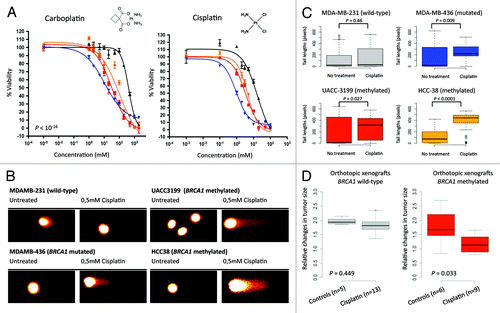
We transferred our experiments from the in vitro assays described above to an in vivo setting in a mouse model. The antitumor activity of cisplatin was evaluated with respect to BRCA1 epigenetic status using UACC3199 (BRCA1 hypermethylated) and MDA-MB-231 (BRCA1 unmethylated) cancer cells xenografted in nude mice. Upon subcutaneous administration of cisplatin, significant tumor growth inhibition over time was observed in the BRCA1 hypermethylated xenografts (p = 0.025), but not in unmethylated cells (p = 0.443). The mice were sacrificed 30 d after the start of the treatment and the tumor size of the xenograft was measured. BRCA1 hypermethylated cells had significantly smaller tumors than the xenografted unmethylated cells (p = 0.033) ().
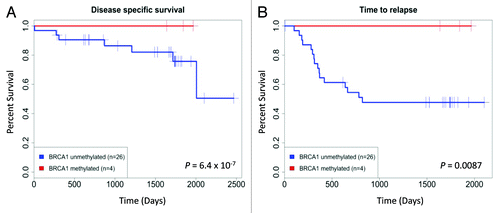
Given the aforementioned in vitro and in vivo findings that human cancer cells with BRCA1-methylation-associated silencing are very sensitive to platin derivatives, we wondered whether the same could be observed in clinical samples. In the clinical context, cisplatin is a chemotherapy drug widely used in the treatment of ovarian cancer, a tumor type in which a significant rate of BRCA1 CpG island hypermethylation has been described.Citation17-Citation22 We therefore assessed whether the presence of BRCA1 promoter CpG island hypermethylation, detected by pyrosequencing, was a predictive marker of response to cisplatin in ovarian cancer patients treated with this drug. The study of a well characterized clinical cohort of serous epithelial ovarian tumors [FIGO Stages: I (n = 7), II (n = 3), III (n = 18) and IV (n = 2)], all of which were treated with cisplatin, showed that BRCA1 methylation was observed in 13% (4 of 30) of the cases. The BRCA1 hypermethylated ovarian tumors corresponded to FIGO stages I (n = 2) and II (n = 2). Most importantly, BRCA1 epigenetic inactivation was associated with a significantly longer time to relapse (Cox regression, log-rank, p = 6.40E-007) and improved overall survival (Cox regression, log-rank, p = 0.009) (). Thus, the clinical data resemble the aforementioned cell culture and xenograft results that suggest an increased chemosensitivity of BRCA1 hypermethylated tumors to platinum-derived drugs.
Figure 3. BRCA1 hypermethylation proves to be a predictor of good response to chemotherapy with cisplatin in ovarian cancer patients. (A) BRCA1 hypermethylation in patients with ovarian cancer is associated with longer time to relapse. (B) BRCA1 hypermethylation in patients with ovarian cancer is associated with improved disease-specific survival.
One of the “holy grails” of current medical oncology is personalized cancer treatment. The oncologist would like to have information available that pinpoints a particular molecular Achilles’ heel in a given patient that indicates the usefulness of a particular drug. To date, this approach has been most successful for treating hematological malignancies, but progress with solid tumors, such as breast, colon and lung tumors has also been made. A number of studies in ovarian tumorsCitation5,Citation6 support the hypothesis that inherited genetic defects in BRCA1/BRCA2 render these neoplasms more sensitive to platinum-based regimens. Herein, using the BRCA1 epigenetic defect, we have broadened these observations to include sporadic tumors, which make up the vast majority of cases attended by medical practitioners. Our results support the inclusion of BRCA1 promoter CpG island hypermethylation in biomarker panels assessing the clinical efficacy of platinum-based chemotherapy.
Acknowledgments
Supported by Grants SAF2011–22803, Lilly Foundation, Botin Foundation, Dr. Josef Steiner Cancer Research Foundation, Fundación Sandra Ibarra de Solidaridad frente al Cáncer, Junta de Barcelona of the Asociación Española Contra el Cáncer (AECC), and the Health and Science Departments of the Catalan Government (Generalitat de Catalunya). M.E. is an ICREA Research Professor.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- Roukos DH, Briasoulis E. Individualized preventive and therapeutic management of hereditary breast ovarian cancer syndrome. Nat Clin Pract Oncol 2007; 4:578 - 90; http://dx.doi.org/10.1038/ncponc0930; PMID: 17898808
- Roy R, Chun J, Powell SN. BRCA1 and BRCA2: different roles in a common pathway of genome protection. Nat Rev Cancer 2012; 12:68 - 78; http://dx.doi.org/10.1038/nrc3181; PMID: 22193408
- Byrski T, Gronwald J, Huzarski T, Grzybowska E, Budryk M, Stawicka M, et al. Pathologic complete response rates in young women with BRCA1-positive breast cancers after neoadjuvant chemotherapy. J Clin Oncol 2010; 28:375 - 9; http://dx.doi.org/10.1200/JCO.2008.20.7019; PMID: 20008645
- Silver DP, Richardson AL, Eklund AC, Wang ZC, Szallasi Z, Li Q, et al. Efficacy of neoadjuvant Cisplatin in triple-negative breast cancer. J Clin Oncol 2010; 28:1145 - 53; http://dx.doi.org/10.1200/JCO.2009.22.4725; PMID: 20100965
- Yang D, Khan S, Sun Y, Hess K, Shmulevich I, Sood AK, et al. Association of BRCA1 and BRCA2 mutations with survival, chemotherapy sensitivity, and gene mutator phenotype in patients with ovarian cancer. JAMA 2011; 306:1557 - 65; http://dx.doi.org/10.1001/jama.2011.1456; PMID: 21990299
- Bolton KL, Chenevix-Trench G, Goh C, Sadetzki S, Ramus SJ, Karlan BY, et al, EMBRACE, kConFab Investigators, Cancer Genome Atlas Research Network. Association between BRCA1 and BRCA2 mutations and survival in women with invasive epithelial ovarian cancer. JAMA 2012; 307:382 - 90; http://dx.doi.org/10.1001/jama.2012.20; PMID: 22274685
- Kang J, D’Andrea AD, Kozono D. A DNA repair pathway-focused score for prediction of outcomes in ovarian cancer treated with platinum-based chemotherapy. J Natl Cancer Inst 2012; 104:670 - 81; http://dx.doi.org/10.1093/jnci/djs177; PMID: 22505474
- Fong PC, Boss DS, Yap TA, Tutt A, Wu P, Mergui-Roelvink M, et al. Inhibition of poly(ADP-ribose) polymerase in tumors from BRCA mutation carriers. N Engl J Med 2009; 361:123 - 34; http://dx.doi.org/10.1056/NEJMoa0900212; PMID: 19553641
- Fong PC, Yap TA, Boss DS, Carden CP, Mergui-Roelvink M, Gourley C, et al. Poly(ADP)-ribose polymerase inhibition: frequent durable responses in BRCA carrier ovarian cancer correlating with platinum-free interval. J Clin Oncol 2010; 28:2512 - 9; http://dx.doi.org/10.1200/JCO.2009.26.9589; PMID: 20406929
- Audeh MW, Carmichael J, Penson RT, Friedlander M, Powell B, Bell-McGuinn KM, et al. Oral poly(ADP-ribose) polymerase inhibitor olaparib in patients with BRCA1 or BRCA2 mutations and recurrent ovarian cancer: a proof-of-concept trial. Lancet 2010; 376:245 - 51; http://dx.doi.org/10.1016/S0140-6736(10)60893-8; PMID: 20609468
- Kaye SB, Lubinski J, Matulonis U, Ang JE, Gourley C, Karlan BY, et al. Phase II, open-label, randomized, multicenter study comparing the efficacy and safety of olaparib, a poly (ADP-ribose) polymerase inhibitor, and pegylated liposomal doxorubicin in patients with BRCA1 or BRCA2 mutations and recurrent ovarian cancer. J Clin Oncol 2012; 30:372 - 9; http://dx.doi.org/10.1200/JCO.2011.36.9215; PMID: 22203755
- Ledermann J, Harter P, Gourley C, Friedlander M, Vergote I, Rustin G, et al. Olaparib maintenance therapy in platinum-sensitive relapsed ovarian cancer. N Engl J Med 2012; 366:1382 - 92; http://dx.doi.org/10.1056/NEJMoa1105535; PMID: 22452356
- Heyn H, Esteller M. DNA methylation profiling in the clinic: applications and challenges. Nat Rev Genet 2012; 13:679 - 92; http://dx.doi.org/10.1038/nrg3270; PMID: 22945394
- Esteller M, Garcia-Foncillas J, Andion E, Goodman SN, Hidalgo OF, Vanaclocha V, et al. Inactivation of the DNA-repair gene MGMT and the clinical response of gliomas to alkylating agents. N Engl J Med 2000; 343:1350 - 4; http://dx.doi.org/10.1056/NEJM200011093431901; PMID: 11070098
- Hegi ME, Diserens AC, Gorlia T, Hamou MF, de Tribolet N, Weller M, et al. MGMT gene silencing and benefit from temozolomide in glioblastoma. N Engl J Med 2005; 352:997 - 1003; http://dx.doi.org/10.1056/NEJMoa043331; PMID: 15758010
- Bleeker FE, Molenaar RJ, Leenstra S. Recent advances in the molecular understanding of glioblastoma. J Neurooncol 2012; 108:11 - 27; http://dx.doi.org/10.1007/s11060-011-0793-0; PMID: 22270850
- Catteau A, Harris WH, Xu CF, Solomon E. Methylation of the BRCA1 promoter region in sporadic breast and ovarian cancer: correlation with disease characteristics. Oncogene 1999; 18:1957 - 65; http://dx.doi.org/10.1038/sj.onc.1202509; PMID: 10208417
- Magdinier F, Ribieras S, Lenoir GM, Frappart L, Dante R. Down-regulation of BRCA1 in human sporadic breast cancer; analysis of DNA methylation patterns of the putative promoter region. Oncogene 1998; 17:3169 - 76; http://dx.doi.org/10.1038/sj.onc.1202248; PMID: 9872332
- Rice JC, Massey-Brown KS, Futscher BW. Aberrant methylation of the BRCA1 CpG island promoter is associated with decreased BRCA1 mRNA in sporadic breast cancer cells. Oncogene 1998; 17:1807 - 12; http://dx.doi.org/10.1038/sj.onc.1202086; PMID: 9778046
- Esteller M, Silva JM, Dominguez G, Bonilla F, Matias-Guiu X, Lerma E, et al. Promoter hypermethylation and BRCA1 inactivation in sporadic breast and ovarian tumors. J Natl Cancer Inst 2000; 92:564 - 9; http://dx.doi.org/10.1093/jnci/92.7.564; PMID: 10749912
- Esteller M, Fraga MF, Guo M, Garcia-Foncillas J, Hedenfalk I, Godwin AK, et al. DNA methylation patterns in hereditary human cancers mimic sporadic tumorigenesis. Hum Mol Genet 2001; 10:3001 - 7; http://dx.doi.org/10.1093/hmg/10.26.3001; PMID: 11751682
- Grushko TA, Dignam JJ, Das S, Blackwood AM, Perou CM, Ridderstråle KK, et al. MYC is amplified in BRCA1-associated breast cancers. Clin Cancer Res 2004; 10:499 - 507; http://dx.doi.org/10.1158/1078-0432.CCR-0976-03; PMID: 14760071
- Geisler JP, Hatterman-Zogg MA, Rathe JA, Buller RE. Frequency of BRCA1 dysfunction in ovarian cancer. J Natl Cancer Inst 2002; 94:61 - 7; http://dx.doi.org/10.1093/jnci/94.1.61; PMID: 11773283
- Ibanez de Caceres I, Battagli C, Esteller M, Herman JG, Dulaimi E, Edelson MI, et al. Tumor cell-specific BRCA1 and RASSF1A hypermethylation in serum, plasma, and peritoneal fluid from ovarian cancer patients. Cancer Res 2004; 64:6476 - 81; http://dx.doi.org/10.1158/0008-5472.CAN-04-1529; PMID: 15374957
- Cancer Genome Atlas Research Network. Integrated genomic analyses of ovarian carcinoma. Nature 2011; 474:609 - 15; http://dx.doi.org/10.1038/nature10166; PMID: 21720365
- Hedenfalk I, Duggan D, Chen Y, Radmacher M, Bittner M, Simon R, et al. Gene-expression profiles in hereditary breast cancer. N Engl J Med 2001; 344:539 - 48; http://dx.doi.org/10.1056/NEJM200102223440801; PMID: 11207349
- Veeck J, Ropero S, Setien F, Gonzalez-Suarez E, Osorio A, Benitez J, et al. BRCA1 CpG island hypermethylation predicts sensitivity to poly(adenosine diphosphate)-ribose polymerase inhibitors. J Clin Oncol 2010; 28:e563 - 4, author reply e565-6; http://dx.doi.org/10.1200/JCO.2010.30.1010; PMID: 20679605
- Drew Y, Mulligan EA, Vong WT, Thomas HD, Kahn S, Kyle S, et al. Therapeutic potential of poly(ADP-ribose) polymerase inhibitor AG014699 in human cancers with mutated or methylated BRCA1 or BRCA2. J Natl Cancer Inst 2011; 103:334 - 46; http://dx.doi.org/10.1093/jnci/djq509; PMID: 21183737