Abstract
Microrchidia (MORC) is a highly conserved nuclear protein superfamily with widespread domain architectures that intimately link MORCs with signaling-dependent chromatin remodeling and epigenetic regulation. Accumulating structural and biochemical evidence has shed new light on the mechanistic action and emerging role of MORCs as epigenetic regulators in diverse nuclear processes. In this Point of View, we focus on discussing recent advances in our understanding of the unique domain architectures of MORC family of chromatin remodelers and their potential contribution to epigenetic control of DNA template-dependent processes such as transcription and DNA damage response. Given that the deregulation of MORCs has been linked with human cancer and other diseases, further efforts to uncover the structure and function of MORCs may ultimately lead to the development of new approaches to intersect with the functionality of MORC family of chromatin remodeling proteins to correct associated pathogenesis.
Introduction
Microrchidia (MORC) is a relatively uncharacterized, highly conserved nuclear protein family from prokaryotic to eukaryotic cells.Citation1,Citation2 To date, there are five members of the MORC family in humans, namely, MORC1 (also called MORC, ZCW6 or CT33), MORC2 (ZCW3, ZCWCC1, KIAA0852 or AC004542.C22.1), MORC3 (ZCW5, ZCWCC3, NXP2 or KIAA0136), MORC4 (ZCW4, ZCWCC2, FLJ11565 or dJ75H8.2) and the divergent SMCHD1 (structural maintenance of chromosomes flexible hinge domain containing 1) (KIAA0650) with chromosome condensation protein SMC-type hinge domains in addition to the MORC module (www.genecards.org).Citation1,Citation3-Citation5
Emerging evidence shows that members of the MORC protein superfamily exert tissue-specific expression patterns with a wide range of biological functions (). In this context, it was initially thought that MORC1 is primarily expressed in male germ cells and regulates mammalian germ cell development and meiosis.Citation3,Citation6 However, recent studies provided the evidence that MORC1 is frequently expressed in multiple myeloma cellsCitation7 and is frequently mutated in primary and metastatic estrogen receptor-positive lobular breast cancers.Citation8 In contrast, MORC2 is ubiquitously expressed in human cells and tissues.Citation4,Citation16,Citation17 Gene expression profiling studies revealed that the levels of MORC2 expression are upregulated in breast cancer tissues and in situ carcinomas, as compared with adjacent normal breast tissues,Citation9 and associated with the recurrence of triple-negative breast cancer.Citation10
Table 1. Expression patterns of MORCs
Promyelocytic leukemia-nuclear body (PML-NB) is a dynamic subnuclear macromolecular structure formed by PML and Sp100 proteins and has been implicated in the regulation of diverse cellular functions, including transcription, DNA repair and tumor suppression.Citation18-Citation20 Interestingly, MORC3 has been shown to localize on PML-NBs and to induce p53-dependent premature senescence through regulating p53 activation and localization into PML-NBs.Citation11 More recent studies demonstrated a two-step mechanism involved in the co-localization of MORC3 with PML-NBs.Citation12 In this context, MORC3 functions as a “molecular clamp” through the ATPase cycle to form MORC3 nuclear domains in a PML-independent manner. MORC3 also associates with PML via its sumoylation modification.Citation12 Given that many of the proteins that accumulate in PML-NBs are putative epigenetic factors such as histone methyltransferases, histone deacetylases or DNA methyltransferases,Citation21 it is conceivable that MORC3 is likely to be an epigenetic regulator in diverse biological processes. In addition, it is interesting to note that the expression of MORC3 is significantly altered in normal peripheral blood leukocytes following treatment with chemotherapy agents, but its suspected role in chemotherapy response remains to be determined.Citation13 In contrast, MORC4 mRNA is widely expressed at low levels in normal tissues, with highest expression levels in placenta and testis.Citation14 Recently, MORC4 has been identified as a potential lymphoma biomarker, as it is highly expressed in about 66% diffuse large B-cell lymphomas patients and in multiple B-cell lymphoma-derived cell lines.Citation14 However, the functional role and the underlying mechanism of MORC4 in the development and progression of lymphoma remain unexplored.
SMCHD1 belongs to the structural maintenance of chromosome (SMC) gene superfamily that plays fundamental roles in higher-order chromosome organization and dynamics.Citation22,Citation23 Protein sequence analysis revealed that mouse Smchd1 shares 86% sequence identity with its human homolog.Citation24 SMCHD1 has been recently identified as a causal genetic determinant of facioscapulohumeral dystrophy type 2Citation15 as well as a potential tumor suppressor in human cancers.Citation24 It is noteworthy to mention that SMCHD1, like MORC3,Citation12 is a potential substrate of sumoylation,Citation25 but whether and how the sumoylation modification plays a role in the regulation of SMCHD1 function under physiological and pathophysiological conditions remains to be tested.
The accumulating structural and biochemical evidence has provided new perspective about the mechanism of action and functions of MORCs. In particular, MORCs are becoming increasingly recognized as new epigenetic regulators of fundamental biological processes.Citation4,Citation26 In the following sections, we discuss recent advances in our understanding of the domain architectures of MORCs, the intriguing roles of MORCs in epigenetic regulation of transcription and DNA damage response, and the potential connection between these two processes.
Domain Architectures of MORCs
GHKL-type ATPase domain
One characteristic of the MORC protein family is the presence at the N-terminus of a highly conserved GHKL (DNA gyrase, hsp90, histidine kinase and DNA mismatch repair enzyme MutL) domain combined to a carboxyl-terminal S5 domainCitation1-Citation3,Citation27 (). The GHKL domain contains conserved ATP binding motifs, and the S5 domain usually provides a conserved basic residue that functions similarly to the arginine or lysine finger observed in various phosphorylation reaction.Citation1 Thus, the GHKL+ S5 domain constitutes an active GHKL-type ATPase module that is involved in ATP binding and hydrolysis and plays crucial roles in DNA metabolism and signaling transduction.Citation1,Citation2,Citation28 In contrast, the ATPase subunits of the well-characterized, four classes of chromatin remodeling complexes including SWI/SNF (SWItch/sucrose nonfermentable), ISWI (imitation switch), CHD (chromodomain, helicase, DNA binding) and INO80 (inositol requiring 80) contain a common Snf2-like ATPase domainCitation29,Citation30 (). Interestingly, the GHKL-type ATPase is also found in other prokaryotic and eukaryotic chromatin-related proteins, such as heat shock protein 90,Citation31 DNA repair proteins of the MutL family,Citation1,Citation32,Citation33 the ATPase subunits of the topoisomerases,Citation34 SMCHD family member GMI1 (gamma-irradiation and mitomycin C induced 1) in Arabidopsis thalianaCitation35 and defective in meristem silencing 11 (DMS11) in Arabidopsis thaliana.Citation36 It has been shown that the MORC ATPase is involved in gene silencing in Arabidopsis thalianaCitation26,Citation36 and in the regulation of chromatin architecture in response to DNA damage signals in human cellsCitation4 (). Thus, the GHKL-type ATPase module is becoming a new player in epigenetic regulation of transcription and DNA damage response in plants and mammals.
Figure 1. Comparison of the domain architecture between the MORC family (A) and the well-characterized four classes of chromatin remodeling complexes (B). (A) Members of the MORC family contain a conserved GHKL-type ATPase domain at their N-terminus, a PHD-X/ZF-CW domain in their midst and varied coiled-coil domains. In addition, MORC2 protein contains a chromo-like domain at its carboxy-terminus. (B) The ATPase subunits of the four classes of chromatin remodeling complexes including SWI/SNF (SWItch/sucrose non-fermentable), ISWI (imitation switch), CHD (chromodomain, helicase, DNA binding) and INO80 (inositol requiring 80) contain a common Snf2-like ATPase domain and other functional domains as indicated.Citation29,Citation30
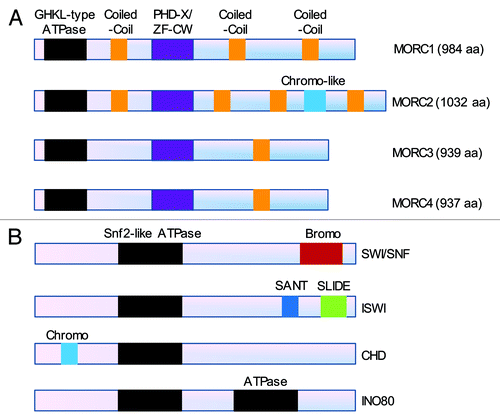
Table 2. Domain architectures of MORCs
PHD-X/ZF-CW domain
In addition to the GHKL-ATPase domain, different members of the MORC family show fusions to other domains that play a critical role in recognizing different epigenetic marks on DNA.Citation4,Citation40 In this context, another widespread domain of the MORC proteins is a carboxyl-terminal PHD-X/ZF-CW domainCitation40 (). The ZF-CW domain is a motif of about 60 amino acids comprising at least four cysteine (C) and two tryptophan (W) residues that is frequently found in proteins involved in epigenetic regulation.Citation37,Citation38,Citation40 This domain is an N-terminally truncated version of the zinc-binding PHD (plant homeodomain) finger domain that lacks the first and third metal-chelating dyads of the binuclear treble clef fold.Citation37,Citation41 Despite this structural modification, it retains the histone H3 binding interface and predominantly functions as a methylated H3K4 binding domain, similar to the conventional PHD domainCitation2,Citation37-Citation39,Citation41 ().
In addition to MORCs, the ZF-CW domain is also present in other chromatin-related factors (www.uniprot.org/uniprot), such as Arabidopsis thaliana histone-lysine N-methyltransferase ASHH2,Citation42 HSI2 (high-level expression of sugar-inducible gene 2), HSL1 (HSI2-like 1),Citation43,Citation44 methyl-CpG-binding domain-containing protein 1 (AtMBD1), AtMBD2, AtMBD3, AtMBD4, AtMBD12,Citation45,Citation46 human lysine-specific histone demethylase 1B (KDM1B) (also known as LSD2 or AOF1),Citation47,Citation48 zinc finger CW-type PWWP domain protein 1 protein 1 (ZCWPW1)Citation37 and ZCWPW2. Recent studies have shown that HSL1 interacts with histone deacetylase 19 (HDA19) through its ZF-CW domain and recruits HDA19 to repress the expression of seed maturation genes in seedlings.Citation43 In addition, KDM1B contains a unique N-terminal ZF-CW domain that is not present in KDM1A (also known as LSD1 or AOF2)Citation47,Citation49 and is required for its demethylase activity and transcriptional repression activity.Citation49,Citation50 Furthermore, structural and biochemical evidence has shown that the ZF-CW domains of ASHH2, ZCWPW1, MORC3, MORC4 and HSI2 proteins are novel histone recognition modules with specifically for methylated histone H3 lysine 4,Citation37-Citation39 but their binding preference for the histone H3 methylation status may vary in different proteins.Citation38 These observations suggest that the ZF-CW domain is involved in chromatin regulation through the recognition of epigenetic signals.Citation37
Chomo-like domain
The chromo-like domain is a widely distributed sequence motif comprising about 40–50 amino acids and is involved in the epigenetic regulation of heterochromatin function and euchromatic gene expression.Citation51-Citation53 Computational analysis of MORC2 sequence, through a hidden Markov model search using the JACKHMMER program (hmmer.janelia.org) for further conserved domains that might have previously eluded detection, recognized a novel chromo-like domain that recovers the first chromo-like domain of histone methylase SETDB1Citation54 (). All elements comprising the core SH3-like fold of the chromo-like domains are conserved in the version of the domain found in MORC2,Citation41 suggesting that it is likely to bind to lysines in histones. Chromo-like domain is widely present in various chromatin-associated proteins in humans (www.uniprot.org/uniprot), such as the chromodomain-helicase-DNA-binding (CHD) protein family members (CHD1–9), chromobox protein homolog (CBX) family members (CBX1–8), histone acetyltransferases KAT5 and KAT8, histone methyltransferases SUV39H1 and SUV39H2, SWI/SNF complex subunits SMARCC1 and SMARCC2, AT-rich interactive domain-containing protein 4A (ARID4A), ARID4B, chromodomain Y-like protein 2 (CDYL2), testis-specific chromodomain protein Y 1 (CDY1), CDY2, M-phase phosphoprotein 8 (MPHOSPH8), male-specific lethal 3 homolog (MSL3) and mortality factor 4 like 1(MORF4L1).Citation51-Citation53 These proteins have been widely documented with respect to dominant functions in chromatin remodeling and the regulation of gene expression in health and disease.Citation51-Citation53 However, the function and mechanism of action of the chromo-like domain in MORC2 protein have not yet been elucidated.
Coiled-coil domain
The coiled-coil domain typically consists of two to five α-helices wrapped around each other into super-helical structures and is found in about 10% of all protein sequences.Citation55-Citation57 Accumulating evidence has suggested that the coiled-coil domain is an important structural determinant for the regulation of protein-protein and protein-DNA interaction,Citation57-Citation66 protein stability,Citation67,Citation68 protein functional activation,Citation65,Citation69 subcellular localization,Citation65,Citation70-Citation72 gene transcription,Citation73,Citation74 DNA damage responseCitation75-Citation78 and signaling transduction.Citation66,Citation79-Citation83 Interestingly, MORC1 and MORC2 contain the predicted three-stranded coiled-coil domain that is absent in both MORC3 and MORC4 proteins.Citation40 Instead, MORC3 and MORC4 have an additional two-stranded coiled-coil motif near their carboxyl-terminusCitation40(). However, the implication of these conserved coiled-coil domains in emerging MORC functions remains to be investigated.
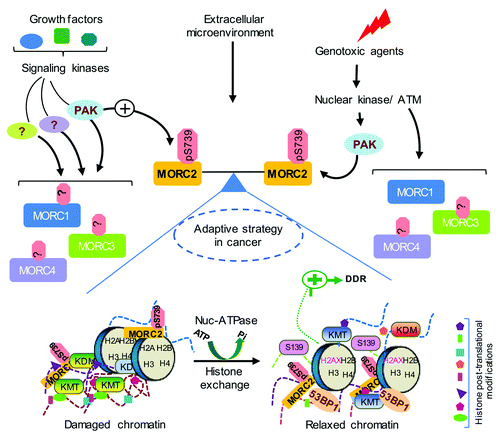
Collectively, recent structural and biochemical studies have defined MORCs as potential epigenetic regulators of many basic biological processes. It is becoming increasingly clear that crosstalk between different structural domains plays an important role in the regulation of protein functions. One case in point is the human chromatin remodeler CHD4, whose PHD and chromo domains associate with its ATPase motif and regulate its ATPase activity.Citation84 However, it remains unclear whether and how these structural domains of MORCs act in concert to govern their biological functions in response to extracellular and intracellular stimuli. Interestingly, our recent study revealed that signaling-dependent phosphorylation of MORC2 at lysine 739 that localizes between the PHD-X/ZF-CW domain and the chromo-like domain controls its ATPase activity responsible for chromatin remodeling in response to DNA damage signals, but he mechanastic detail remains elusive.Citation4
Functions of MORCs
MORCs and transcription
Accumulating evidence suggests that MORCs play a conserved role in transcription (). In this context, two Arabidopsis genes, AtMORC1 (also known as CRT1) and AtMORC6 (also known as DMS11) are involved in heterochromatin condensation and gene silencing in Arabidopsis thaliana.Citation26 Arabidopsis thaliana CRT1 (compromised for recognition of Turnip Crinkle virus) is a prototypic eukaryotic member of the MORC superfamily with a conserved GHKL-ATPase motif.Citation27,Citation89 It has been recently shown that CRT1 binds DNA, exhibits endonuclease activity, and has important nuclear functions during immune response.Citation88 In contrast, DMS11 is a GHKL-type ATPase that is also involved in RNA-directed DNA methylation in Arabidopsis thaliana.Citation36 In this context, DMS11 interacts with DMS3, which lacks ATPase motif, and provides the missing ATPase function for DMS3, and consequently, cooperate in the RNA-directed DNA methylation pathway to promote transcriptional repression.Citation36
Table 3. Functions of MORCs in transcription and DNA damage response
Similarly, MORC2 has been shown to repress carbonic anhydrase IX (CAIX) gene expression in gastric cancer cells through interacting with and recruiting histone deacetylase 4 (HDAC4) onto the CAIX promoter.Citation16 In addition, MORC3 is a nuclear matrix-associated protein with RNA binding activity, raising the possibility that MORC3 might have a novel function in nuclear RNA metabolism.Citation5 In support of this notion, MORC3 associates with SUMO-2 and is required for transcriptional repression.Citation85
Consistent with the above observations, SMCHD1 has a critical role in epigenetic gene silencing.Citation22 In this context, SMCHD1 localizes to the inactive X chromosome and has a critical role in the maintenance of X inactivation and the hypermethylation of CpG islands (CGIs).Citation22 A recent study further pointed out that inactive X chromosome CpG island methylation occurs via either Smchd1-dependent or independent pathway.Citation86 In this context, a subset of CGIs is methylated at a relatively fast rate following the onset of X inactivation that is independent of the chromosomal protein Smchd1 in many cases. In contrast, methylation of other CGIs proceeds relatively slowly and requires Smchd1.Citation86 Smchd1 has been recently identified as an epigenetic modifier of the D4Z4 metastable epiallele and as a causal genetic determinant of facioscapulohumeral dystrophy type 2.Citation15 Interestingly, a recent study identified Smchd1 as a tumor suppressor in oncogene-driven models.Citation24 Lack of Smchd1 is not inherently oncogenic, but loss of Smchd1 accelerates Eμ-Myc-driven B-cell lymphomas by deregulating expression of a subset of Polycomb-repressive complex 2 and mixed lineage leukemia fusion protein target genes.Citation24 However, lack of Smchd1 does not affect overall telomere length in mice.Citation90 Interestingly, DMS3, a protein with homology to Smchd1 in Arabidopsis thaliana, has been shown to be required for RNA-mediated DNA methylation.Citation87 In support of this notion, the GHKL ATPase DMS11 physically interacts and cooperates with DMS3 in the RNA-mediated DNA methylation pathway to promote transcriptional repression.Citation36
MORC and DNA damage response (DDR)
Given their nuclear localization signals, MORCs have been predicted to be a component of the DNA damage response signaling network in addition to its role in transcriptionCitation3 (). In support of this notion, CRT1 (AtMORC1) has been shown to enhance tolerance to DNA-damage agent mitomycin C, indicating that CRT1 is involved in DNA damage repair.Citation88 Our recent study discovered that MORC2 is activated via phosphorylation by extracellular DNA damage signals and facilitates an ATPase-dependent chromatin remodeling following DNA damage induction, thus promoting DNA double-strand break (DSB) repair and cell survivalCitation4 ().
Figure 2. Proposed model for the physiologic role of MORCs. MORCs control a variety of cellular and physiological functions in response to growth factor signaling, extracellular microenvironment, and genotoxic stress. Posttranslational modification of MORCs by upstream kinases might modulate their cellular functions. PAK1 phosphorylates MORC2 at serine 739 in response to both genotoxic stress and growth factor signaling and directs an effective DDR by an ATP-dependent chromatin remodeling event. Domain architecture of MORCs will provide essential insights into their specific and non-redundant roles in epigenetic regulation via recognition of various posttranslational modifications of histones or functional interactions with histone modifying enzymes. MORCs are deregulated in a variety of cancers and possibly function as a critical balance for efficient crosstalk between growth factor signaling and DDR. Collectively, these will contribute to adaptive survival strategies and therapy resistance often observed in cancer. DDR, DNA damage response.
The histone variant H2AX is a key chromatin component in cellular responses to DSBs and, in particular, the phosphorylation of H2AX (termed γH2AX) by the phosphatidylinositol 3-kinase-related kinases (PIKKs) ataxia telangiectasia mutated (ATM), ataxia telangiectasia and Rad3 related (ATR) and DNA-dependent protein kinase (DNA-PK) is a specific and key coordinator of DDR signaling.Citation91-Citation94 Our laboratory has recently demonstrated that MORC2 promotes the induction of γH2AX following DNA damage, but has no any effect on the expression and activation of the ATM, ATR, DNA-PK protein kinases.Citation4 Similarly, the well-characterized SWI/SNF chromatin remodeling complex regulates DNA damage-induced γH2AX induction, but does not affect the expression of the above mentioned PIKK kinases, or their activation and/or recruitment to DSBs.Citation95 Thus, a functional crosstalk between the MORC2-containing complex and the SWI/SNF complex in recognition and signaling of DSBs in the context of chromatin remains to be investigated in the future.Citation4 In addition, it has been well documented that the INO80 chromatin remodeling complex is recruited to DSB sites through its interaction with γH2AX.Citation96,Citation97 Thus, it will be interesting to examine whether MORC2-containing chromatin remodeling complex is recruited to DNA damage sites and whether this recruitment depends on its interaction with γH2AX.
Another interesting finding is that MORC2 exerts a histone exchange activity that enables to replace the nucleosomal H2AZ-H2B dimer with the canonical H2A-H2B dimer in a phosphorylation- and ATPase-dependent manner.Citation4 However, the functional implication of MORC2-mediated histone exchange in DSB repair or other basic biological processes remains largely unknown. Interestingly, a recent study revealed that the p400 remodeling ATPase utilizes its ATPase activity to exchange histone H2AZ onto nucleosomes at DSBs.Citation98 Moreover, the H2AZ exchange promotes specific patterns of histone modification and regulates the loading of the DSB repair complexes at DSBs, thus contributing to DSB repair.Citation98 Similarly, the INO80 chromatin remodeling complex also has a histone-exchange activity that exchanges nucleosomal H2AZ-H2B with free H2A-H2B dimers to promote genome stability.Citation99 In S. cerevisiae, the SWR1 chromatin remodeling complex catalyzes ATP-dependent exchange of nucleosomal histone H2A for H2AZ (S. cerevisiae Htz1) that is closely linked to the tight regulation of transcription and genetic stability.Citation100,Citation101 Given these interestingly observations, it will be important to examine whether and how the MORCs and other chromatin remodeling complexes coordinately reorganize chromatin architecture in response to DNA damage and other stresses.
In addition, given that MORC3 localizes on PML-NBs,Citation11,Citation12 which are intrinsic elements of the cellular response to DNA damage,Citation12,Citation102 it would be exciting to examine whether MORC3 is implicated in DNA damage response in the future. In addition, GMI1, a GHKL-type ATPase domain containing protein in Arabidopsis and the homologs of mammalian SMCHD1, is induced by γ-irradiation in an ataxia telangiectasia mutated (ATM)-dependent manner and involved in the repair of DNA double-strand breaks, presumably, by homologous recombination.Citation35 However, it remains unknown whether the mammalian SMCHD1 plays a role in DNA repair in addition to its known function in X-inactivation.Citation35
MORCs: Moving Forward
The evolutionary contextual and gene neighborhood studies on prokaryotic MORCs and their eukaryotic relatives as well as recently emerging structural and biochemical evidence have defined MORCs as new players in epigenetic regulators of genome.Citation1,Citation2,Citation4,Citation26,Citation37,Citation38 Despite these great strides a detailed comprehensive analysis to delineate distinct non-redundant roles of these proteins in various cellular responses await functional and biochemical evaluation. To this end, a recent report by the authors has provided a first-of-its-kind, detailed biochemical analysis supporting the role and chromatin remodeling functions of MORC2 protein for optimal cellular response to genotoxic stress,Citation4 and thereby established a functional link between this novel nucleosome-dependent ATPase and genome-surveillance mechanisms (). Intriguingly, MORC2’s ATPase-dependent chromatin remodeling function is dictated by upstream kinase dependent signaling in response to DNA-damage response and growth factor signaling.Citation4 In light of our current knowledge, it is tempting to speculate that MORC2 may be a key epigenetic contributor in adaptive strategies employed by tumor cells for survival and therapy resistance. At the cellular level this can be achieved by efficiently integrating growth factor signaling with DNA repair processes. Mounting evidence suggests that MORCs play crucial roles in various cellular processes integral to maintain cellular homeostasis.
A closer analysis of MORC proteins and sequence comparison studies suggest the presence of conserved novel histone-recognition or sensing modules (). Chromatin remodelers like MORC2 function to alter nucleosomal structure of chromatin (). We predict that MORC proteins are likely to remodel nucleosomes with specific single or multivalent histone modifications utilizing unique histone-reading and -editing functions. We also suggest that MORCs may recognize similar or different single or combinatorial histone modifications and this, in turn, may impact the recognition patterns due to specific posttranslational modifications by upstream signaling events. Answers to these questions would fuel and shape future research to understand the physiological roles of these novel chromatin remodelers. Recent advancement in next-generation sequencing techniques like ChIP-Seq technology with robust supportive bioinformatics tools have provided novel insights to understand genome-wide recruitment patterns of various chromatin-interacting proteins at very high resolution. In this context, ChIP-Seq recruitment patterns of MORCs under defined experimental conditions combined with RNA sequencing of human cells with defined modulation of expression and functions of MORC2 have revealed a genome-wide footprint of MORC2 and its impact on gene expression and provided invaluable clues about “yet-to-be-discovered” targets and functions of MORC2 in eukaryotic cells (SSN, DQL and RK; unpublished data). Given that deregulation of MORCs has been linked with human cancers and other diseases,Citation3,Citation7,Citation8,Citation10,Citation14,Citation16,Citation17,Citation88 further studies may lead to better understanding of the components influenced by MORCs that could be targeted to reverse MORC-associated pathologic conditions. In closing, our understanding of the structural insights and physiological roles of eukaryotic MORCs and the contribution of the epigenome in the phenotypic outcome are likely to be merely the tip of the iceberg. Ongoing studies in our and other laboratories are addressing some of the emerging questions outlined here, with an aim to reveal molecular insights of the role of MORCs in transcription, chromatin looping, histone sensing and DNA repair.
Acknowledgments
We apologize to all of our colleagues whose work has not been cited here due to space limitations. The genome-wide MORC discovery work is supported by the McCormick Proteomic and Genomic Center of the George Washington University, and work describing MORC2 in DNA damage response is supported by National Institutes of Health grant CA139573 (to R.K.).
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Notes
† These authors contributed equally to this work.
References
- Iyer LM, Abhiman S, Aravind L. MutL homologs in restriction-modification systems and the origin of eukaryotic MORC ATPases. Biol Direct 2008; 3:8; http://dx.doi.org/10.1186/1745-6150-3-8; PMID: 18346280
- Iyer LM, Anantharaman V, Wolf MY, Aravind L. Comparative genomics of transcription factors and chromatin proteins in parasitic protists and other eukaryotes. Int J Parasitol 2008; 38:1 - 31; http://dx.doi.org/10.1016/j.ijpara.2007.07.018; PMID: 17949725
- Inoue N, Hess KD, Moreadith RW, Richardson LL, Handel MA, Watson ML, et al. New gene family defined by MORC, a nuclear protein required for mouse spermatogenesis. Hum Mol Genet 1999; 8:1201 - 7; http://dx.doi.org/10.1093/hmg/8.7.1201; PMID: 10369865
- Li DQ, Nair SS, Ohshiro K, Kumar A, Nair VS, Pakala SB, et al. MORC2 signaling integrates phosphorylation-dependent, ATPase-coupled chromatin remodeling during the DNA damage response. Cell Rep 2012; 2:1657 - 69; http://dx.doi.org/10.1016/j.celrep.2012.11.018; PMID: 23260667
- Kimura Y, Sakai F, Nakano O, Kisaki O, Sugimoto H, Sawamura T, et al. The newly identified human nuclear protein NXP-2 possesses three distinct domains, the nuclear matrix-binding, RNA-binding, and coiled-coil domains. J Biol Chem 2002; 277:20611 - 7; http://dx.doi.org/10.1074/jbc.M201440200; PMID: 11927593
- Watson ML, Zinn AR, Inoue N, Hess KD, Cobb J, Handel MA, et al. Identification of morc (microrchidia), a mutation that results in arrest of spermatogenesis at an early meiotic stage in the mouse. Proc Natl Acad Sci U S A 1998; 95:14361 - 6; http://dx.doi.org/10.1073/pnas.95.24.14361; PMID: 9826705
- Condomines M, Hose D, Raynaud P, Hundemer M, De Vos J, Baudard M, et al. Cancer/testis genes in multiple myeloma: expression patterns and prognosis value determined by microarray analysis. J Immunol 2007; 178:3307 - 15; PMID: 17312182
- Shah SP, Morin RD, Khattra J, Prentice L, Pugh T, Burleigh A, et al. Mutational evolution in a lobular breast tumour profiled at single nucleotide resolution. Nature 2009; 461:809 - 13; http://dx.doi.org/10.1038/nature08489; PMID: 19812674
- Tripathi A, King C, de la Morenas A, Perry VK, Burke B, Antoine GA, et al. Gene expression abnormalities in histologically normal breast epithelium of breast cancer patients. Int J Cancer 2008; 122:1557 - 66; http://dx.doi.org/10.1002/ijc.23267; PMID: 18058819
- Chen LH, Kuo WH, Tsai MH, Chen PC, Hsiao CK, Chuang EY, et al. Identification of prognostic genes for recurrent risk prediction in triple negative breast cancer patients in Taiwan. PLoS One 2011; 6:e28222; http://dx.doi.org/10.1371/journal.pone.0028222; PMID: 22140552
- Takahashi K, Yoshida N, Murakami N, Kawata K, Ishizaki H, Tanaka-Okamoto M, et al. Dynamic regulation of p53 subnuclear localization and senescence by MORC3. Mol Biol Cell 2007; 18:1701 - 9; http://dx.doi.org/10.1091/mbc.E06-08-0747; PMID: 17332504
- Mimura Y, Takahashi K, Kawata K, Akazawa T, Inoue N. Two-step colocalization of MORC3 with PML nuclear bodies. J Cell Sci 2010; 123:2014 - 24; http://dx.doi.org/10.1242/jcs.063586; PMID: 20501696
- González-Fernández R, Morales M, Avila J, Martín-Vasallo P. Changes in leukocyte gene expression profiles induced by antineoplastic chemotherapy. Oncol Lett 2012; 3:1341 - 9; PMID: 22783446
- Liggins AP, Cooper CD, Lawrie CH, Brown PJ, Collins GP, Hatton CS, et al. MORC4, a novel member of the MORC family, is highly expressed in a subset of diffuse large B-cell lymphomas. Br J Haematol 2007; 138:479 - 86; http://dx.doi.org/10.1111/j.1365-2141.2007.06680.x; PMID: 17608765
- Lemmers RJ, Tawil R, Petek LM, Balog J, Block GJ, Santen GW, et al. Digenic inheritance of an SMCHD1 mutation and an FSHD-permissive D4Z4 allele causes facioscapulohumeral muscular dystrophy type 2. Nat Genet 2012; 44:1370 - 4; http://dx.doi.org/10.1038/ng.2454; PMID: 23143600
- Shao Y, Li Y, Zhang J, Liu D, Liu F, Zhao Y, et al. Involvement of histone deacetylation in MORC2-mediated down-regulation of carbonic anhydrase IX. Nucleic Acids Res 2010; 38:2813 - 24; http://dx.doi.org/10.1093/nar/gkq006; PMID: 20110259
- Wang GL, Wang CY, Cai XZ, Chen W, Wang XH, Li F. Identification and expression analysis of a novel CW-type zinc finger protein MORC2 in cancer cells. Anat Rec (Hoboken) 2010; 293:1002 - 9; http://dx.doi.org/10.1002/ar.21119; PMID: 20225202
- Bernardi R, Pandolfi PP. Structure, dynamics and functions of promyelocytic leukaemia nuclear bodies. Nat Rev Mol Cell Biol 2007; 8:1006 - 16; http://dx.doi.org/10.1038/nrm2277; PMID: 17928811
- Draskovic I, Arnoult N, Steiner V, Bacchetti S, Lomonte P, Londoño-Vallejo A. Probing PML body function in ALT cells reveals spatiotemporal requirements for telomere recombination. Proc Natl Acad Sci U S A 2009; 106:15726 - 31; http://dx.doi.org/10.1073/pnas.0907689106; PMID: 19717459
- Lang M, Jegou T, Chung I, Richter K, Münch S, Udvarhelyi A, et al. Three-dimensional organization of promyelocytic leukemia nuclear bodies. J Cell Sci 2010; 123:392 - 400; http://dx.doi.org/10.1242/jcs.053496; PMID: 20130140
- Torok D, Ching RW, Bazett-Jones DP. PML nuclear bodies as sites of epigenetic regulation. Front Biosci 2009; 14:1325 - 36; http://dx.doi.org/10.2741/3311; PMID: 19273133
- Blewitt ME, Gendrel AV, Pang Z, Sparrow DB, Whitelaw N, Craig JM, et al. SmcHD1, containing a structural-maintenance-of-chromosomes hinge domain, has a critical role in X inactivation. Nat Genet 2008; 40:663 - 9; http://dx.doi.org/10.1038/ng.142; PMID: 18425126
- Losada A, Hirano T. Dynamic molecular linkers of the genome: the first decade of SMC proteins. Genes Dev 2005; 19:1269 - 87; http://dx.doi.org/10.1101/gad.1320505; PMID: 15937217
- Leong HS, Chen K, Hu Y, Lee S, Corbin J, Pakusch M, et al. Epigenetic regulator Smchd1 functions as a tumor suppressor. Cancer Res 2013; 73:1591 - 9; http://dx.doi.org/10.1158/0008-5472.CAN-12-3019; PMID: 23269277
- Tirard M, Hsiao HH, Nikolov M, Urlaub H, Melchior F, Brose N. In vivo localization and identification of SUMOylated proteins in the brain of His6-HA-SUMO1 knock-in mice. Proc Natl Acad Sci U S A 2012; 109:21122 - 7; http://dx.doi.org/10.1073/pnas.1215366110; PMID: 23213215
- Moissiard G, Cokus SJ, Cary J, Feng S, Billi AC, Stroud H, et al. MORC family ATPases required for heterochromatin condensation and gene silencing. Science 2012; 336:1448 - 51; http://dx.doi.org/10.1126/science.1221472; PMID: 22555433
- Dutta R, Inouye M. GHKL, an emergent ATPase/kinase superfamily. Trends Biochem Sci 2000; 25:24 - 8; http://dx.doi.org/10.1016/S0968-0004(99)01503-0; PMID: 10637609
- Hu X, Machius M, Yang W. Monovalent cation dependence and preference of GHKL ATPases and kinases. FEBS Lett 2003; 544:268 - 73; http://dx.doi.org/10.1016/S0014-5793(03)00519-2; PMID: 12782329
- Vignali M, Hassan AH, Neely KE, Workman JL. ATP-dependent chromatin-remodeling complexes. Mol Cell Biol 2000; 20:1899 - 910; http://dx.doi.org/10.1128/MCB.20.6.1899-1910.2000; PMID: 10688638
- Clapier CR, Cairns BR. The biology of chromatin remodeling complexes. Annu Rev Biochem 2009; 78:273 - 304; http://dx.doi.org/10.1146/annurev.biochem.77.062706.153223; PMID: 19355820
- Meyer P, Prodromou C, Hu B, Vaughan C, Roe SM, Panaretou B, et al. Structural and functional analysis of the middle segment of hsp90: implications for ATP hydrolysis and client protein and cochaperone interactions. Mol Cell 2003; 11:647 - 58; http://dx.doi.org/10.1016/S1097-2765(03)00065-0; PMID: 12667448
- Ban C, Junop M, Yang W. Transformation of MutL by ATP binding and hydrolysis: a switch in DNA mismatch repair. Cell 1999; 97:85 - 97; http://dx.doi.org/10.1016/S0092-8674(00)80717-5; PMID: 10199405
- Räschle M, Dufner P, Marra G, Jiricny J. Mutations within the hMLH1 and hPMS2 subunits of the human MutLalpha mismatch repair factor affect its ATPase activity, but not its ability to interact with hMutSalpha. J Biol Chem 2002; 277:21810 - 20; http://dx.doi.org/10.1074/jbc.M108787200; PMID: 11948175
- Corbett KD, Berger JM. Structural dissection of ATP turnover in the prototypical GHL ATPase TopoVI. Structure 2005; 13:873 - 82; http://dx.doi.org/10.1016/j.str.2005.03.013; PMID: 15939019
- Böhmdorfer G, Schleiffer A, Brunmeir R, Ferscha S, Nizhynska V, Kozák J, et al. GMI1, a structural-maintenance-of-chromosomes-hinge domain-containing protein, is involved in somatic homologous recombination in Arabidopsis. Plant J 2011; 67:420 - 33; http://dx.doi.org/10.1111/j.1365-313X.2011.04604.x; PMID: 21481027
- Lorković ZJ, Naumann U, Matzke AJ, Matzke M. Involvement of a GHKL ATPase in RNA-directed DNA methylation in Arabidopsis thaliana. Curr Biol 2012; 22:933 - 8; http://dx.doi.org/10.1016/j.cub.2012.03.061; PMID: 22560611
- He F, Umehara T, Saito K, Harada T, Watanabe S, Yabuki T, et al. Structural insight into the zinc finger CW domain as a histone modification reader. Structure 2010; 18:1127 - 39; http://dx.doi.org/10.1016/j.str.2010.06.012; PMID: 20826339
- Hoppmann V, Thorstensen T, Kristiansen PE, Veiseth SV, Rahman MA, Finne K, et al. The CW domain, a new histone recognition module in chromatin proteins. EMBO J 2011; 30:1939 - 52; http://dx.doi.org/10.1038/emboj.2011.108; PMID: 21522130
- Li X, Foley EA, Molloy KR, Li Y, Chait BT, Kapoor TM. Quantitative chemical proteomics approach to identify post-translational modification-mediated protein-protein interactions. J Am Chem Soc 2012; 134:1982 - 5; http://dx.doi.org/10.1021/ja210528v; PMID: 22239320
- Perry J, Zhao Y. The CW domain, a structural module shared amongst vertebrates, vertebrate-infecting parasites and higher plants. Trends Biochem Sci 2003; 28:576 - 80; http://dx.doi.org/10.1016/j.tibs.2003.09.007; PMID: 14607086
- Aravind L, Abhiman S, Iyer LM. Natural history of the eukaryotic chromatin protein methylation system. Prog Mol Biol Transl Sci 2011; 101:105 - 76; http://dx.doi.org/10.1016/B978-0-12-387685-0.00004-4; PMID: 21507350
- Zhao Z, Yu Y, Meyer D, Wu C, Shen WH. Prevention of early flowering by expression of FLOWERING LOCUS C requires methylation of histone H3 K36. Nat Cell Biol 2005; 7:1256 - 60; http://dx.doi.org/10.1038/ncb1329; PMID: 16299497
- Zhou Y, Tan B, Luo M, Li Y, Liu C, Chen C, et al. HISTONE DEACETYLASE19 interacts with HSL1 and participates in the repression of seed maturation genes in Arabidopsis seedlings. Plant Cell 2013; 25:134 - 48; http://dx.doi.org/10.1105/tpc.112.096313; PMID: 23362207
- Suzuki M, Wang HH, McCarty DR. Repression of the LEAFY COTYLEDON 1/B3 regulatory network in plant embryo development by VP1/ABSCISIC ACID INSENSITIVE 3-LIKE B3 genes. Plant Physiol 2007; 143:902 - 11; http://dx.doi.org/10.1104/pp.106.092320; PMID: 17158584
- Zemach A, Grafi G. Methyl-CpG-binding domain proteins in plants: interpreters of DNA methylation. Trends Plant Sci 2007; 12:80 - 5; http://dx.doi.org/10.1016/j.tplants.2006.12.004; PMID: 17208509
- Ito M, Koike A, Koizumi N, Sano H. Methylated DNA-binding proteins from Arabidopsis. Plant Physiol 2003; 133:1747 - 54; http://dx.doi.org/10.1104/pp.103.026708; PMID: 14605234
- Karytinos A, Forneris F, Profumo A, Ciossani G, Battaglioli E, Binda C, et al. A novel mammalian flavin-dependent histone demethylase. J Biol Chem 2009; 284:17775 - 82; http://dx.doi.org/10.1074/jbc.M109.003087; PMID: 19407342
- Ciccone DN, Su H, Hevi S, Gay F, Lei H, Bajko J, et al. KDM1B is a histone H3K4 demethylase required to establish maternal genomic imprints. Nature 2009; 461:415 - 8; http://dx.doi.org/10.1038/nature08315; PMID: 19727073
- Yang Z, Jiang J, Stewart DM, Qi S, Yamane K, Li J, et al. AOF1 is a histone H3K4 demethylase possessing demethylase activity-independent repression function. Cell Res 2010; 20:276 - 87; http://dx.doi.org/10.1038/cr.2010.12; PMID: 20101264
- Zhang Q, Qi S, Xu M, Yu L, Tao Y, Deng Z, et al. Structure-function analysis reveals a novel mechanism for regulation of histone demethylase LSD2/AOF1/KDM1b. Cell Res 2013; 23:225 - 41; http://dx.doi.org/10.1038/cr.2012.177; PMID: 23266887
- Cavalli G, Paro R. Chromo-domain proteins: linking chromatin structure to epigenetic regulation. Curr Opin Cell Biol 1998; 10:354 - 60; http://dx.doi.org/10.1016/S0955-0674(98)80011-2; PMID: 9640536
- Tajul-Arifin K, Teasdale R, Ravasi T, Hume DA, Mattick JS, RIKEN GER Group, GSL Members. Identification and analysis of chromodomain-containing proteins encoded in the mouse transcriptome. Genome Res 2003; 13:6B 1416 - 29; http://dx.doi.org/10.1101/gr.1015703; PMID: 12819141
- Jones DO, Cowell IG, Singh PB. Mammalian chromodomain proteins: their role in genome organisation and expression. Bioessays 2000; 22:124 - 37; http://dx.doi.org/10.1002/(SICI)1521-1878(200002)22:2<124::AID-BIES4>3.0.CO;2-E; PMID: 10655032
- Ceol CJ, Houvras Y, Jane-Valbuena J, Bilodeau S, Orlando DA, Battisti V, et al. The histone methyltransferase SETDB1 is recurrently amplified in melanoma and accelerates its onset. Nature 2011; 471:513 - 7; http://dx.doi.org/10.1038/nature09806; PMID: 21430779
- Mason JM, Arndt KM. Coiled coil domains: stability, specificity, and biological implications. Chembiochem 2004; 5:170 - 6; http://dx.doi.org/10.1002/cbic.200300781; PMID: 14760737
- McFarlane AA, Orriss GL, Stetefeld J. The use of coiled-coil proteins in drug delivery systems. Eur J Pharmacol 2009; 625:101 - 7; http://dx.doi.org/10.1016/j.ejphar.2009.05.034; PMID: 19835864
- Lupas AN, Gruber M. The structure of α-helical coiled coils. Adv Protein Chem 2005; 70:37 - 78; http://dx.doi.org/10.1016/S0065-3233(05)70003-6; PMID: 15837513
- Li X, He L, Che KH, Funderburk SF, Pan L, Pan N, et al. Imperfect interface of Beclin1 coiled-coil domain regulates homodimer and heterodimer formation with Atg14L and UVRAG. Nat Commun 2012; 3:662; http://dx.doi.org/10.1038/ncomms1648; PMID: 22314358
- Burkhard P, Stetefeld J, Strelkov SV. Coiled coils: a highly versatile protein folding motif. Trends Cell Biol 2001; 11:82 - 8; http://dx.doi.org/10.1016/S0962-8924(00)01898-5; PMID: 11166216
- Cheng P, Yang Y, Heintzen C, Liu Y. Coiled-coil domain-mediated FRQ-FRQ interaction is essential for its circadian clock function in Neurospora. EMBO J 2001; 20:101 - 8; http://dx.doi.org/10.1093/emboj/20.1.101; PMID: 11226160
- Peng H, Begg GE, Schultz DC, Friedman JR, Jensen DE, Speicher DW, et al. Reconstitution of the KRAB-KAP-1 repressor complex: a model system for defining the molecular anatomy of RING-B box-coiled-coil domain-mediated protein-protein interactions. J Mol Biol 2000; 295:1139 - 62; http://dx.doi.org/10.1006/jmbi.1999.3402; PMID: 10653693
- Hurme R, Berndt KD, Namork E, Rhen M. DNA binding exerted by a bacterial gene regulator with an extensive coiled-coil domain. J Biol Chem 1996; 271:12626 - 31; http://dx.doi.org/10.1074/jbc.271.21.12626; PMID: 8647874
- Nikolay R, Wiederkehr T, Rist W, Kramer G, Mayer MP, Bukau B. Dimerization of the human E3 ligase CHIP via a coiled-coil domain is essential for its activity. J Biol Chem 2004; 279:2673 - 8; http://dx.doi.org/10.1074/jbc.M311112200; PMID: 14610072
- Parachoniak CA, Park M. Distinct recruitment of Eps15 via Its coiled-coil domain is required for efficient down-regulation of the met receptor tyrosine kinase. J Biol Chem 2009; 284:8382 - 94; http://dx.doi.org/10.1074/jbc.M807607200; PMID: 19109251
- Tanner MJ, Hanel W, Gaffen SL, Lin X. CARMA1 coiled-coil domain is involved in the oligomerization and subcellular localization of CARMA1 and is required for T cell receptor-induced NF-kappaB activation. J Biol Chem 2007; 282:17141 - 7; http://dx.doi.org/10.1074/jbc.M700169200; PMID: 17428801
- Tao W, Malone CL, Ault AD, Deschenes RJ, Fassler JS. A cytoplasmic coiled-coil domain is required for histidine kinase activity of the yeast osmosensor, SLN1. Mol Microbiol 2002; 43:459 - 73; http://dx.doi.org/10.1046/j.1365-2958.2002.02757.x; PMID: 11985722
- Fanelli M, Fantozzi A, De Luca P, Caprodossi S, Matsuzawa S, Lazar MA, et al. The coiled-coil domain is the structural determinant for mammalian homologues of Drosophila Sina-mediated degradation of promyelocytic leukemia protein and other tripartite motif proteins by the proteasome. J Biol Chem 2004; 279:5374 - 9; http://dx.doi.org/10.1074/jbc.M306407200; PMID: 14645235
- Yang K, Zhu J, Sun S, Tang Y, Zhang B, Diao L, et al. The coiled-coil domain of TRAF6 is essential for its auto-ubiquitination. Biochem Biophys Res Commun 2004; 324:432 - 9; http://dx.doi.org/10.1016/j.bbrc.2004.09.070; PMID: 15465037
- Zhang T, Kee WH, Seow KT, Fung W, Cao X. The coiled-coil domain of Stat3 is essential for its SH2 domain-mediated receptor binding and subsequent activation induced by epidermal growth factor and interleukin-6. Mol Cell Biol 2000; 20:7132 - 9; http://dx.doi.org/10.1128/MCB.20.19.7132-7139.2000; PMID: 10982829
- Ma J, Zhang T, Novotny-Diermayr V, Tan AL, Cao X. A novel sequence in the coiled-coil domain of Stat3 essential for its nuclear translocation. J Biol Chem 2003; 278:29252 - 60; http://dx.doi.org/10.1074/jbc.M304196200; PMID: 12746441
- Raiborg C, Bremnes B, Mehlum A, Gillooly DJ, D’Arrigo A, Stang E, et al. FYVE and coiled-coil domains determine the specific localisation of Hrs to early endosomes. J Cell Sci 2001; 114:2255 - 63; PMID: 11493665
- Begitt A, Meyer T, van Rossum M, Vinkemeier U. Nucleocytoplasmic translocation of Stat1 is regulated by a leucine-rich export signal in the coiled-coil domain. Proc Natl Acad Sci U S A 2000; 97:10418 - 23; http://dx.doi.org/10.1073/pnas.190318397; PMID: 10973496
- Blazek D, Barboric M, Kohoutek J, Oven I, Peterlin BM. Oligomerization of HEXIM1 via 7SK snRNA and coiled-coil region directs the inhibition of P-TEFb. Nucleic Acids Res 2005; 33:7000 - 10; http://dx.doi.org/10.1093/nar/gki997; PMID: 16377779
- Watanabe M, Yanagi Y, Masuhiro Y, Yano T, Yoshikawa H, Yanagisawa J, et al. A putative tumor suppressor, TSG101, acts as a transcriptional suppressor through its coiled-coil domain. Biochem Biophys Res Commun 1998; 245:900 - 5; http://dx.doi.org/10.1006/bbrc.1998.8547; PMID: 9588212
- Buisson R, Masson JY. PALB2 self-interaction controls homologous recombination. Nucleic Acids Res 2012; 40:10312 - 23; http://dx.doi.org/10.1093/nar/gks807; PMID: 22941656
- Hohl M, Kwon Y, Galván SM, Xue X, Tous C, Aguilera A, et al. The Rad50 coiled-coil domain is indispensable for Mre11 complex functions. Nat Struct Mol Biol 2011; 18:1124 - 31; http://dx.doi.org/10.1038/nsmb.2116; PMID: 21892167
- Itakura E, Sawada I, Matsuura A. Dimerization of the ATRIP protein through the coiled-coil motif and its implication to the maintenance of stalled replication forks. Mol Biol Cell 2005; 16:5551 - 62; http://dx.doi.org/10.1091/mbc.E05-05-0427; PMID: 16176973
- Ball HL, Cortez D. ATRIP oligomerization is required for ATR-dependent checkpoint signaling. J Biol Chem 2005; 280:31390 - 6; http://dx.doi.org/10.1074/jbc.M504961200; PMID: 16027118
- Bar M, Sharfman M, Schuster S, Avni A. The coiled-coil domain of EHD2 mediates inhibition of LeEix2 endocytosis and signaling. PLoS One 2009; 4:e7973; http://dx.doi.org/10.1371/journal.pone.0007973; PMID: 19936242
- Rairdan GJ, Collier SM, Sacco MA, Baldwin TT, Boettrich T, Moffett P. The coiled-coil and nucleotide binding domains of the Potato Rx disease resistance protein function in pathogen recognition and signaling. Plant Cell 2008; 20:739 - 51; http://dx.doi.org/10.1105/tpc.107.056036; PMID: 18344282
- He Y, Wertheim JA, Xu L, Miller JP, Karnell FG, Choi JK, et al. The coiled-coil domain and Tyr177 of bcr are required to induce a murine chronic myelogenous leukemia-like disease by bcr/abl. Blood 2002; 99:2957 - 68; http://dx.doi.org/10.1182/blood.V99.8.2957; PMID: 11929787
- Cheng HY, Schiavone AP, Smithgall TE. A point mutation in the N-terminal coiled-coil domain releases c-Fes tyrosine kinase activity and survival signaling in myeloid leukemia cells. Mol Cell Biol 2001; 21:6170 - 80; http://dx.doi.org/10.1128/MCB.21.18.6170-6180.2001; PMID: 11509660
- Zhang X, Subrahmanyam R, Wong R, Gross AW, Ren R. The NH(2)-terminal coiled-coil domain and tyrosine 177 play important roles in induction of a myeloproliferative disease in mice by Bcr-Abl. Mol Cell Biol 2001; 21:840 - 53; http://dx.doi.org/10.1128/MCB.21.3.840-853.2001; PMID: 11154271
- Watson AA, Mahajan P, Mertens HD, Deery MJ, Zhang W, Pham P, et al. The PHD and chromo domains regulate the ATPase activity of the human chromatin remodeler CHD4. J Mol Biol 2012; 422:3 - 17; http://dx.doi.org/10.1016/j.jmb.2012.04.031; PMID: 22575888
- Rosendorff A, Sakakibara S, Lu S, Kieff E, Xuan Y, DiBacco A, et al. NXP-2 association with SUMO-2 depends on lysines required for transcriptional repression. Proc Natl Acad Sci U S A 2006; 103:5308 - 13; http://dx.doi.org/10.1073/pnas.0601066103; PMID: 16567619
- Gendrel AV, Apedaile A, Coker H, Termanis A, Zvetkova I, Godwin J, et al. Smchd1-dependent and -independent pathways determine developmental dynamics of CpG island methylation on the inactive X chromosome. Dev Cell 2012; 23:265 - 79; http://dx.doi.org/10.1016/j.devcel.2012.06.011; PMID: 22841499
- Kanno T, Bucher E, Daxinger L, Huettel B, Böhmdorfer G, Gregor W, et al. A structural-maintenance-of-chromosomes hinge domain-containing protein is required for RNA-directed DNA methylation. Nat Genet 2008; 40:670 - 5; http://dx.doi.org/10.1038/ng.119; PMID: 18425128
- Kang HG, Hyong WC, von Einem S, Manosalva P, Ehlers K, Liu PP, et al. CRT1 is a nuclear-translocated MORC endonuclease that participates in multiple levels of plant immunity. Nat Commun 2012; 3:1297; http://dx.doi.org/10.1038/ncomms2279; PMID: 23250427
- Kang HG, Kuhl JC, Kachroo P, Klessig DF. CRT1, an Arabidopsis ATPase that interacts with diverse resistance proteins and modulates disease resistance to turnip crinkle virus. Cell Host Microbe 2008; 3:48 - 57; http://dx.doi.org/10.1016/j.chom.2007.11.006; PMID: 18191794
- Roberts AR, Blewitt ME, Youngson NA, Whitelaw E, Chong S. Reduced dosage of the modifiers of epigenetic reprogramming Dnmt1, Dnmt3L, SmcHD1 and Foxo3a has no detectable effect on mouse telomere length in vivo. Chromosoma 2011; 120:377 - 85; http://dx.doi.org/10.1007/s00412-011-0318-9; PMID: 21553025
- Stiff T, O’Driscoll M, Rief N, Iwabuchi K, Löbrich M, Jeggo PA. ATM and DNA-PK function redundantly to phosphorylate H2AX after exposure to ionizing radiation. Cancer Res 2004; 64:2390 - 6; http://dx.doi.org/10.1158/0008-5472.CAN-03-3207; PMID: 15059890
- Ward IM, Chen J. Histone H2AX is phosphorylated in an ATR-dependent manner in response to replicational stress. J Biol Chem 2001; 276:47759 - 62; PMID: 11673449
- Kinner A, Wu W, Staudt C, Iliakis G. Gamma-H2AX in recognition and signaling of DNA double-strand breaks in the context of chromatin. Nucleic Acids Res 2008; 36:5678 - 94; http://dx.doi.org/10.1093/nar/gkn550; PMID: 18772227
- Rogakou EP, Pilch DR, Orr AH, Ivanova VS, Bonner WM. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J Biol Chem 1998; 273:5858 - 68; http://dx.doi.org/10.1074/jbc.273.10.5858; PMID: 9488723
- Park JH, Park EJ, Lee HS, Kim SJ, Hur SK, Imbalzano AN, et al. Mammalian SWI/SNF complexes facilitate DNA double-strand break repair by promoting gamma-H2AX induction. EMBO J 2006; 25:3986 - 97; http://dx.doi.org/10.1038/sj.emboj.7601291; PMID: 16932743
- van Attikum H, Fritsch O, Hohn B, Gasser SM. Recruitment of the INO80 complex by H2A phosphorylation links ATP-dependent chromatin remodeling with DNA double-strand break repair. Cell 2004; 119:777 - 88; http://dx.doi.org/10.1016/j.cell.2004.11.033; PMID: 15607975
- Morrison AJ, Highland J, Krogan NJ, Arbel-Eden A, Greenblatt JF, Haber JE, et al. INO80 and gamma-H2AX interaction links ATP-dependent chromatin remodeling to DNA damage repair. Cell 2004; 119:767 - 75; http://dx.doi.org/10.1016/j.cell.2004.11.037; PMID: 15607974
- Xu Y, Ayrapetov MK, Xu C, Gursoy-Yuzugullu O, Hu Y, Price BD. Histone H2A.Z controls a critical chromatin remodeling step required for DNA double-strand break repair. Mol Cell 2012; 48:723 - 33; http://dx.doi.org/10.1016/j.molcel.2012.09.026; PMID: 23122415
- Papamichos-Chronakis M, Watanabe S, Rando OJ, Peterson CL. Global regulation of H2A.Z localization by the INO80 chromatin-remodeling enzyme is essential for genome integrity. Cell 2011; 144:200 - 13; http://dx.doi.org/10.1016/j.cell.2010.12.021; PMID: 21241891
- Mizuguchi G, Shen X, Landry J, Wu WH, Sen S, Wu C. ATP-driven exchange of histone H2AZ variant catalyzed by SWR1 chromatin remodeling complex. Science 2004; 303:343 - 8; http://dx.doi.org/10.1126/science.1090701; PMID: 14645854
- Morillo-Huesca M, Clemente-Ruiz M, Andújar E, Prado F. The SWR1 histone replacement complex causes genetic instability and genome-wide transcription misregulation in the absence of H2A.Z. PLoS One 2010; 5:e12143; http://dx.doi.org/10.1371/journal.pone.0012143; PMID: 20711347
- Dellaire G, Ching RW, Ahmed K, Jalali F, Tse KC, Bristow RG, et al. Promyelocytic leukemia nuclear bodies behave as DNA damage sensors whose response to DNA double-strand breaks is regulated by NBS1 and the kinases ATM, Chk2, and ATR. J Cell Biol 2006; 175:55 - 66; http://dx.doi.org/10.1083/jcb.200604009; PMID: 17030982