Abstract
Tensin3 is a cytoskeletal regulatory protein that inhibits cell motility. Downregulation of the gene encoding Tensin3 (TNS3) in human renal cell carcinoma (RCC) may contribute to cancer cell metastatic behavior. We speculated that epigenetic mechanisms, e.g., gene promoter hypermethylation, might account for TNS3 downregulation. In this study, we identified and validated a TNS3 gene promoter containing a CpG island, and quantified the methylation level within this region in RCC. Using a luciferase reporter assay we demonstrated a functional minimal promoter activity for a 500-bp sequence within the TNS3 CpG island. Pyrosequencing enabled quantitative determination of DNA methylation of each CpG dinucleotide (a total of 43) in the TNS3 gene promoter. Across the entire analyzed CpG stretch, RCC DNA showed a higher methylation level than both non-tumor kidney DNA and normal control DNA. Out of all the CpGs analyzed, two CpG dinucleotides, specifically position 2 and 8, showed the most pronounced increases in methylation levels in tumor samples. Furthermore, CpG-specific higher methylation levels were correlated with lower TNS3 gene expression levels in RCC samples. In addition, pharmacological demethylation treatment of cultured kidney cells caused a 3-fold upregulation of Tensin3 expression. In conclusion, these results reveal a differential methylation pattern in the TNS3 promoter occurring in human RCC, suggesting an epigenetic mechanism for aberrant Tensin downregulation in human kidney cancer.
Introduction
The Tensins are a family of intracellular proteins believed to be involved in cell motility and survival. The protein family consists of four members: Tensin1, Tensin2, Tensin3 and Tensin4 (cten).Citation1 Tensins 1–3 contain N-terminal, phosphatase (PTPase) and C2 domain pairings that are homologous to those in the tumor suppressor lipid phosphatase PTEN.Citation2 Tensin1 has been shown to have actin cross-linking and capping activity in its N-terminal region.Citation3,Citation4 All four Tensins contain a tandem pairing of Src homology 2 (SH2) and phosphotyrosine binding (PTB) domains at their C-termini. The PTB domains of Tensins have been shown to bind to various proteins, including β-integrinsCitation5 and receptor tyrosine kinases.Citation6 Furthermore, both SH2 and the PTB domains of Tensin interact with the tumor suppressor Deleted in Liver Cancer-1 (DLC1), suggesting that Tensin-mediated focal adhesion localization may underpin the mechanism for DLC1 tumor suppressive activity.Citation7 In keeping with such a role, Tensin3 has been shown to inhibit mammary cell migration, potentially through its ability to anchor actin to integrins, thus stabilizing the cell and making it less motile.Citation8 Conversely, Tensin4 was shown to compete with Tensin3 for actin-integrin bridging, being upregulated in breast cancer cells in a concomitant manner to Tensin3 downregulation, and thereby increased cell motility.Citation8 In addition, Tensin2 and 3 also have isoform variants that contain an N-terminal C1 domain. We reported that in Tensin2, possession of the isolated C1 domain appears to function in nuclear localization, where it may be important in pathways such as apoptosis.Citation9
All four Tensins are expressed in the kidney,Citation10,Citation11 where they appear to be required for normal kidney development.Citation11-Citation13 We reported previously that Tensin2 and Tensin3 overexpression in kidney cells had an inhibitory effect on cell proliferation, survival and migration, which was linked to a selective suppression of the Akt signaling pathway.Citation2 Moreover, in renal cell carcinoma (RCC), we observed downregulation of expression of all four Tensin genes.Citation10 These observations therefore implicate one or more of the Tensins, most likely Tensin2 and 3, as potential tumor/metastasis suppressors in the kidney. In the United States, 64,770 new cases of renal cancer were predicted to occur in 2012 causing an estimated 13,570 deaths.Citation14 RCC accounts for approximately 80% of all renal cancers, which can be further subdivided into clear cell RCC (ccRCC-75%), papillary RCC (pRCC-15%) and chromophobe RCC (5%). Approximately 70% of ccRCC have a mutational defect in the von Hippel–Lindau tumor suppressor gene (VHL), while in pRCC there is commonly a defects in the MET proto-oncogene.Citation15 The prognosis for advanced stage disease remains poor and, therefore, there is a need to identify novel, more specific, molecular markers for each RCC type that can direct more specific therapies.
Since the discovery of VHL and its role in RCC, thousands of other genes have been assessed for frequent mutations in RCC; however, none of the identified genes are mutated in more than 15% of the tumors. Nevertheless, more than 60 genes have been shown to be epigenetically dysregulated in RCC.Citation16 An example of an epigenetic modification is DNA methylation, which occurs at CpG dinucleotides. CpG dinucleotides are uncommon in the genome, but they are found in relative high abundance in CpG rich regions (CpG islands). About 60% of genes contain CpG islands in their promoter and exonic regions.Citation17,Citation18 These CpG islands are characteristically unmethylatedCitation18; however, during disease states such as cancer, they can become hypermethylated, causing the downregulation of the related gene.Citation16,Citation19,Citation20
In RCC, 83% of tumors have been shown to have alterations in the VHL gene, including both through methylation and mutation.Citation21 Therefore, gene promoter methylation could be an important epigenetic mechanism for dysregulation of tumor suppressor genes in RCC. In the present study, we have analyzed the methylation status of the promoter for the gene encoding Tensin3 (TNS3), as Tensin3 is the most prominent Tensin family member found in the kidney,Citation11 and its downregulation is most pronounced in human RCCCitation10. Furthermore, IGFBP1 and IGFBP3, two genes that are adjacent to the TNS3 gene (chromosomal location 7p12.3), have both been shown to be epigenetically dysregulated in RCC.Citation22 This, together with the observation that genes in close proximity to one another are often dysregulated together in cancer, e.g., VHL and RASS1A genes in RCC,Citation16 adds support to the hypothesis that Tensin3 may also be regulated in this way.
We report here that there is a difference between human RCCs, non-tumor and normal DNA samples in the methylation status of the TNS3 promoter region, which we identified and functionally validated. Furthermore, pharmacological DNA demethylation treatment of kidney cells in culture resulted in upregulation of Tensin3 expression. Therefore, these results suggest that Tensin3 is epigenetically silenced in human kidney cancer.
Results
Bioinformatics identification of the putative TNS3 promoter and incorporated CpG island
The human Tensin3 gene (TNS3) is located on the short arm of chromosome 7 (7p12.3), on the reverse strand (accession number: ENSG00000136205). Bioinformatics analysis revealed that the gene could potentially give rise to several different transcripts through alternative splicing. We focused on the region surrounding the first exon of one the transcripts, TNS3 012, due to that exon lying immediately downstream of a DNA sequence that displayed the characteristics of a potential promoter (). Moreover, the DNA sequence was also close to the first exons of two other TNS3 gene variants, TNS3 003 and TNS3 001.
Figure 1. Bioinformatics analysis of the human TNS3 gene. The TNS3 gene is located on chromosome 7 at 7p12.3 on the reverse strand, 7:47314752–47622156. An 826-bp CpG island was located upstream of exon 1 encoding one of the alternatively spliced transcripts of Tensin3 (TNS3 012). Closer analysis of the CpG island revealed two stretches with a higher concentration of CpG dinucleotides (Cluster 1 and 2). These clusters were subsequently analyzed for alternative methylation by pyrosequencing. The sequences within them to which PCR primers annealed for the pyrosequencing workflow are indicated in bold.
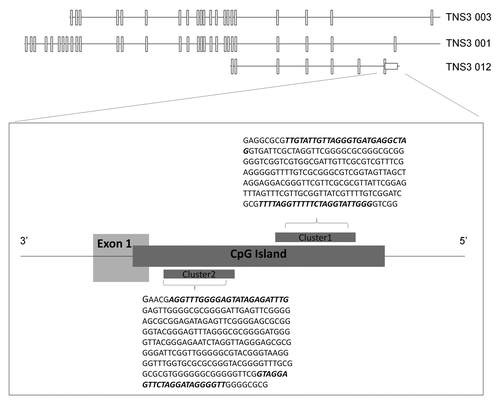
Analysis of the region upstream of this exon 1 showed that it contains an 826-bp CpG island (EMBOSS CpGPlotCitation23), based on the Gardiner-Garden and Frommer criteria for a CpG island: GC content ≥ 50%, Obs/Exp ≥ 0.6 and length of CpG island ≥ 200bpCitation24 (). Within this CpG island, we identified two regions that were particularly enriched in CpG dinucleotides, which we designated Cluster 1 (29 CpGs) and Cluster 2 (14 CpGs). These two clusters were then targeted for PCR amplification after bisulphite conversion, using appropriate primers (MethPrimerCitation25; sequences in Table S1).
Functional promoter activity of the putative TNS3 gene promoter sequence
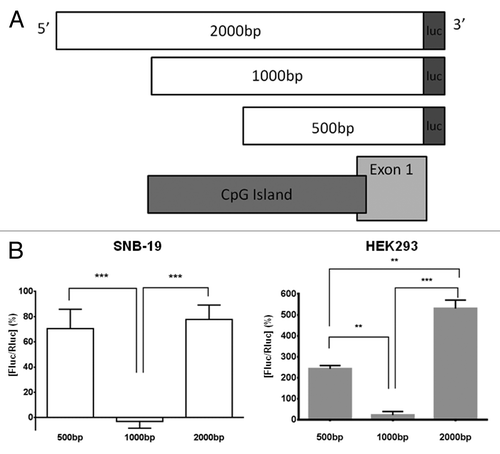
The putative TNS3 gene promoter region we identified was analyzed for functional promoter activity in a luciferase reporter gene assay system. PCR amplified DNA fragments of varying sizes (500 bp, 1,000 bp and 2,000 bp) were cloned into a promoter-less firefly luciferase reporter plasmid (), and transiently expressed in SNB-19 and HEK293 cells. The 500-bp fragment induced luminescence above background levels, reflecting a minimal promoter activity (). Such activity was, however, completely absent with the presence of the longer (1,000-bp) fragment. However, promoter activity was restored with the longest (2,000-bp) fragment to at least the same level or higher (). Furthermore, the same pattern of promoter activity among varying DNA lengths was observed in both human cell lines tested ().
Figure 2. Functional promoter analysis of 2 kb region upstream of exon 1 of the TNS3 gene, TNS3 012 variant. (A) The TNS3 promoter region constructs analyzed for luciferase reporter activity. All three constructs were generated by PCR using a common reverse primer at the end of exon 1 (TNS3 012; ENST00000434451.1) and different forward primers to yield a product of increasing size. These 500-bp, 1,000-bp and 2,000-bp cDNA fragments were then cloned into the pGL4.10 luciferase assay vector; “luc” denotes where each sequence is fused upstream to the firefly luciferase cDNA sequence. As can be seen, the 1,000-bp and 2,000-bp constructs covered the entire CpG island studied here, while the 500-bp fragment does so partially. (B) Luciferase reporter assay of TNS3 promoter function in SNB-19 cells (clear bars) and HEK293 cells (gray bars). Three fragments of this region of different lengths, approximately 500 bp, 1,000 bp and 2 kb, were transfected into SNB-19 and HEK293 cells along with a co-transfection control. The lysates were subsequently analyzed for luminescence as readout of luciferase expression driven by the fragments. The values on the y-axis are the % relative luciferase activity, Fluc/Rluc, derived from the obtained luminescence values with background levels subtracted from them followed by normalization against the transfection control. SNB-19 cells: Bars represent mean luminescence ± SEM (n = 3 experiments); *** denotes p < 0.0001, 1000bp vs. both 500bp and 2000bp fragments. HEK293 cells: Bars represent mean luminescence ± SD (representative of 3 experiments); ***p < 0.001, 1000 bp vs. 2000 bp; **p < 0.01, 500 bp vs. 1000 bp and 500 bp vs. 2000 bp.
Quantitation of TNS3 promoter methylation in cell and RCC patient DNA by pyrosequencing
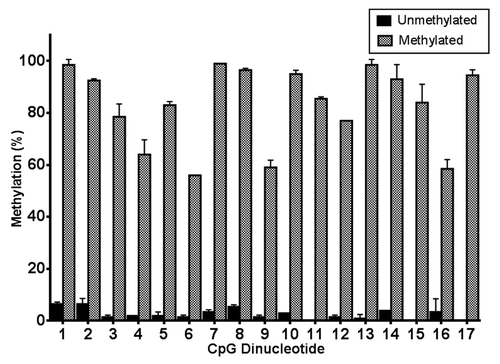
Genomic DNA (gDNA) extracted from HEK293 cells was fully methylated using the bacterial CpG methyltransferase, M.Sssi. This DNA was subsequently used as a methylated control for the pyrosequencing assay, whereas SNB-19 cell gDNA was used as a source of unmethylated DNA, as an unmethylated control for the pyrosequencing assay. We used these DNA samples to validate the accuracy and robustness of the pyrosequencing platform. Pyrosequencing of Cluster 1 region of the TNS3 CpG island revealed the M.Sssi–treated DNA to be highly methylated (). Each individual CpG showed between 50–100% methylation, and the level of methylation across the 17 CpG dinucleotides of Cluster 1 was 83% ± 3.59 (n = 34). In contrast, SNB-19 cell DNA was almost completely unmethylated in the same region, with 0.87% ± 0.29 methylation across the 17 CpGs (n = 34) ().
Figure 3. Validation of pyrosequencing analysis of putative TNS3 promoter in methylated and unmethylated control DNA. Genomic DNA from the HEK293 cell line was treated with the CpG methyltransferase M.Sssi to produce fully methylated control DNA, while the SNB-19 cell DNA served as negative control with negligible methylation. Both gDNA samples underwent pyrosequencing in duplicate; bars represent mean ± SD of methylation level (%) for each CpG dinucleotide within Cluster 1.
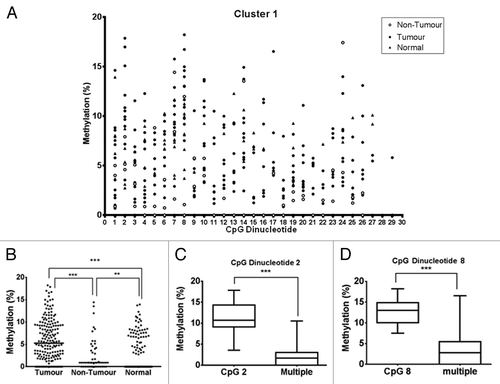
We used the pyrosequencing assay to determine the level of methylation in the TNS3 CpG island in kidney gDNA samples from RCCs and control samples (). In total, 43 CpG dinucleotides were analyzed over the 2 clusters. Overall, we observed significantly higher methylation levels in RCC samples compared with non-tumor kidney samples (; Fig. S1) (Cluster 1, p < 0.001; Cluster 2, p < 0.05). In addition, methylation level in RCC samples was significantly higher compared with normal control samples (Cluster 1: p < 0.0001, RCC vs. normal control; p < 0.01, non-tumor adjacent vs. normal control) (). Furthermore, 1/3 of RCC samples had a median level of methylation over 5% compared with 0% of both non-tumor adjacent and normal control. This overall difference in methylation was largely accounted for by certain individual CpG dinucleotides, in particular, CpG 2 and CpG 8 of Cluster 1 within tumor samples (Kruskal–Wallis H test with post hoc Dunns test). The methylation level in CpG 2 was 10.69%, compared with 1.72% in the significant grouped CpGs (identified in the Kruskal–Wallis H test with post hoc Dunns test) (p < 0.0001, CpG 2 (n = 12) vs. multiple CpGs (n = 80); Mann–Whitney test) (). Also, the methylation level in CpG 8 was 13.01% compared with 2.75% in significant grouped CpGs (p < 0.0001, CpG 8 (n = 12) vs. multiple CpGs (n = 122); Mann–Whitney test) ().
Figure 4. Pyrosequencing analysis of RCC patient and normal control samples. (A) RCC patient kidney gDNA samples were analyzed by pyrosequencing. The Cluster 1 region was sequenced from one PCR product, using two different sequencing primers that enabled coverage of 29 CpG dinucleotides. Each symbol represents the methylation level (%) of an individual sample at each CpG dinucleotide, with filled circles denoting RCC tumor samples (n = 12), open circles denoting adjacent non-tumor kidney samples (n = 3) and filled triangles denoting normal control samples (n = 12). (B) RCC patient samples and normal control samples for the first 17 CpG dinucleotides. ***p < 0.0001, **p < 0.01. (C) and (D) Methylation level of specific CpG dinucleotides within the tumor sample set were compared against each other. Two CpG dinucleotides in Cluster 1, CpG 2 (C) and CpG 8 (D), showed a significantly higher methylation level compared with the remaining CpGs in the sample pool (multiple). Boxes represent mean ± SEM of % methylation. ***p < 0.0001 for CpG 2 vs. multiple and CpG 8 vs. multiple; Mann–Whitney test.
Correlation analysis was performed to evaluate the relationship between the methylation levels on CpGs 2 and 8 and Tensin3 expression level within the same samples. There was significant negative correlation between the Tensin3 mRNA expression in RCC tumors and the methylation level in CpG 2, but not in CpG 8 (). Therefore, increased methylation of CpG 2 was correlated with decreased Tensin3 expression.
Table 1. Correlation between Tensin3 mRNA expression level and methylation level of CpG dinucleotides 2 and 8 in RCC patient DNA
Effect of demethylation of human kidney cell DNA on Tensin3 expression
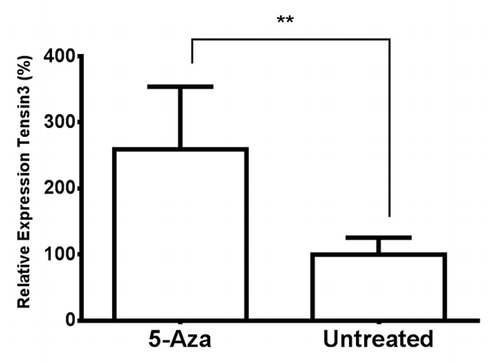
Cultured human kidney cells were treated with the demethylating agent 5-aza in order to determine its effect on Tensin expression. A range of 5-aza concentrations were first tested for their potential cytotoxic effects, and the concentration of 5-aza selected for incubation experiments, 40 µM, was found to be non-toxic to the cultured cells (Fig. S2). qRT-PCR of treated HK-2 cell RNA showed a statistically significant, roughly 3-fold, increase in Tensin3 expression in the 5-aza-treated cells above expression in control, untreated cells (p = 0.0068) (). Interestingly, upregulation of expression by 5-aza treatment was also observed for Tensin2 (p < 0.01) (Fig. S3).
Figure 5. The effect of 5-aza-2′-deoxycytidine treatment on Tensin3 mRNA expression in human kidney cells. HK-2 cells were treated with 40µM 5-aza-2′-deoxycytidine (5-Aza) for 96 h, and RNA extracted from cell lysates underwent qRT-PCR analysis. Tensin3 mRNA amounts were determined using the relative standard curve method, with normalization against the housekeeping gene B2M. Bars represent mean ± SD of gene expression relative to untreated control; **p < 0.01 (n = 3, representative experiment).
Discussion
Dysregulation of members of the Tensin protein family has been observed in various cancer types in a number of studies. We recently reported all Tensins to be downregulated in human RCC.Citation10 Furthermore, Tensins 1–3 have been shown to be downregulated in a number of other cancers,Citation8,Citation10,Citation26-Citation29 while Tensin4 (CTEN), conversely, is mainly upregulated in cancer.Citation30-Citation36 Tensin3 was also shown to be involved in mammary cell migration, where it competed against Tensin4 through mediation of EGF.Citation8 However, since those observations were made, no studies have explored the mechanisms behind Tensin dysregulation in cancer, as a means of understanding the genotypic and phenotypic context within which altered expression takes place. Therefore, in the present study, we investigated a potential epigenetic mechanism for the downregulation of Tensin3 in RCC that we previously observed, focusing on the possibility of aberrant methylation of the TNS3 gene promoter. Our results show that such changes in methylation do take place in RCC and that this is linked to altered Tensin3 expression.
Initial bioinformatics analysis of the TNS3 gene revealed an 826-bp CpG island located immediately upstream of, and running into, exon 1 of one of its various transcripts. Of all the alternative transcript start sites, only this region fulfilled the criteria of being a CpG island. Subsequently, we established and optimized the methodological workflow required in order to analyze in quantitative detail the level of methylation present within this region. By pyrosequencing, we were able to determine the methylation level of up to 43 individual CpG dinucleotides within two CpG clusters that we analyzed separately. Further bioinformatics evidence for this region being a promoter came from the ENCODE database, which revealed a number of histone marks, such as H3K4me1 (enhancer and promoter associated mark), H3K4me3 (promoter associated mark) and H3K27ac (regulatory associated mark), all of which were located upstream of exon 1 of TNS3 003 transcript origin.Citation37 Histone acetylation marks, such as H3K27Ac, are commonly associated with actively transcribed regions, and are usually found around transcription sites, while histone methylation marks are commonly associated with lower transcriptional activity, as methylation appears to stabilize chromatin.Citation38 The precise functions of the H3K4me1and H3K4me3 marks are unknown.Citation39 Furthermore, analysis of the Ensembl genome database identified a CCCTC-binding factor (CTCF) motif also located upstream of the start of exon 1 of TNS3 012. The CTCF factor usually binds to CTCF insulator sequences, which are often located between enhancers and promoters and prevent the enhancer from incorrectly switching on the promoter or activating the wrong promoter.Citation40,Citation41 Also within this region, two other regulatory features exist: a binding site for Ying Yang 1 (YY1), a regulatory transcription factor, and a binding site for E2F1, a transcription factor; both factors have been shown to be involved in cancer dysregulation, e.g., through regulation of tumor suppressor genes or other regulatory genes.Citation42,Citation43 Moreover, bioinformatics software employed by commercial organizations that manufacture gene promoter sequence constructs for functional analysis also identified the same region that we identified as a putative TNS3 gene promoter (www.switchgeargenomics.com).
Having identified the region to study for methylation analysis, we first attempted to experimentally determine a functional promoter activity for the region. The luciferase reporter assay showed a minimal promoter activity to be present in the smallest fragment analyzed, which contains the beginning of the CpG island. Interestingly, the promoter activity was lost when the sequence was extended to a 1,000-bp length, which indicates the presence of a possible repressor element or regulatory feature in the longer stretch, which we have observed in the past for other gene promoters.Citation44 Indeed, bioinformatics analysis showed the presence of a putative regulatory element here, the aforementioned CTCF motif located within this longer sequence. The promoter activity was then restored when testing the largest analyzed fragment (2,000-bp long); in HEK293 cells, this fragment showed greater activity than the 500-bp fragment. Therefore, inclusion of this further stretch could provide further regulatory elements that together produce an overall positive, and perhaps greater, promoter effect, or maybe uncover another distinct gene promoter.
Prior to quantifying the TNS3 promoter methylation in tumor samples from patients with RCC, we validated and optimized our pyrosequencing. To achieve this, we pyrosequenced the CpG island Cluster 1 in genomic DNA samples that either were highly methylated or showed negligible methylation. Our method showed a high degree of robustness and sensitivity and was suitable therefore to be used in subsequent measurement of clinical samples. Analysis of RCC samples showed that overall there was a significantly greater degree of TNS3 CpG island methylation in RCCs samples compared with adjacent non-tumor kidney samples. Moreover, RCC samples showed higher methylation levels (and at a greater significance level) when compared with normal control DNA than when compared with non-tumor samples. Median methylation level was 5.3% in RCCs, 0.9% in non-tumor adjacent cells and 0 in normal control. This observation points to promoter hypermethylation as being a possible cause for the downregulation of Tensin3 mRNA previously reported in RCC.Citation10 Moreover, the specificity and sensitivity of the pyrosequencing approach enabled us to uncover specific locations, at the individual CpG dinucleotide level, where the methylation differences were the greatest. We found that the majority of the higher methylation in the tumor samples were concentrated in two CpG dinucleotides in Cluster 1, specifically CpG 2 and CpG 8. These two dinucleotides were methylated at significantly higher levels than the remaining CpGs around them, indicating that these specific CpGs may play more significant roles in regulating Tensin3 expression. This enhanced role could be due to their close proximity to the regulatory elements CTCF, YY1 and E2F1 and/or proximity to modified histone binding sites around which chromatin remodeling can occur. Additionally, the increase in methylation level seen on CpG 2 was negatively correlated with mRNA expression of Tensin3, i.e., the increase in methylation correlated with a decrease in mRNA expression, indicating that CpG 2 may play a particularly significant role in the epigenetically-driven downregulation of Tensin3 in RCC.
Having measured quantifiable methylation differences between human RCCs, non-tumor and normal DNA, we wished to experimentally determine the effect of pharmacological demethylation of this region on Tensin3 expression. Treatment of human kidney cells with 5-aza caused a significant increase in Tensin3 expression, which demonstrates that Tensin3 expression can be manipulated by exogenously altering the methylation status of the TNS3 promoter. Our qPCR primers/probes recognize all forms of Tensin3 transcript, which makes it clear that Tensin3 expression is low or absent in most cells at the beginning, but that it is most likely that transcripts TNS3 012 and TNS3 001 are the ones that reflect the upregulation after 5-aza treatment. The 5-aza effect could be acting directly on the TNS3 promoter through the inhibition of the methyltransferase DNMT1 or through replacement of methylated cytosines after replication. Alternatively, the effect could be indirect as 5-aza is indiscriminate in targeting hypermethylation;Citation45 therefore, the agent could be upregulating another gene which in turn targets Tensin3 for upregulation. This data, together with evidence that this region in RCC is alternatively methylated vs. adjacent non-tumor and normal control samples, indicates that hypermethylation could play a role in suppressing the expression of Tensin3 as seen in RCC samples. Interestingly, the same upregulating effect of 5-aza was also observed for Tensin2 expression in kidney cells. This was explored due to the fact that the putative TENC1 gene promoter also contains a CpG island, while the genes for Tensin1 and Tensin4 do not.
In conclusion, we have identified a TNS3 gene promoter region through a combination of bioinformatics and experimental approaches. Analysis of the CpG island in this region showed hypermethylation in RCC tumor samples vs. non-tumor samples, suggesting a novel epigenetic mechanism behind the downregulation of Tensin3 in kidney cancer.
Materials and Methods
Cell culture
The human kidney cell lines HK-2 and HEK293 were used for demethylation experiments followed by qRT-PCR.Citation46 The human glioma cell line SNB-19 and HEK293 cells were used in luciferase promoter assays as cells that naturally express Tensin3 and are from the organ of interest, respectivelyCitation47 (Fig. S4). HEK293 cells were also used as a source of genomic DNA for control pyrosequencing experiments. All cells were cultured in complete medium comprised of: Dulbecco’s Modified Eagle Medium (DMEM) supplemented with 10% fetal bovine serum, 20 mM glutamine, 100 U/ml penicillin and 100 μg/ml streptomycin at 37 °C in a humidified atmosphere of 5% CO2 in air.
TNS3 gene promoter activity in luciferase reporter assay
The sequence of the putative TNS3 promoter region, amplified from SNB-19 gDNA, was cloned, in fragments of decreasing length, into the firefly luciferase reporter plasmid pGL4.10 (Promega). Primer sequences for the molecular cloning are provided in Table S2. SNB-19 and HEK293 cells were selected as hosts for the luciferase reporter assay. SNB-19 cells were verified as expressing endogenous Tensin3 naturally, as determined by qRT-PCR and western blot (Fig. S4). Cells were seeded into a 24-well plate in complete medium and allowed to adhere overnight. After this, they were transfected with the reporter plasmids containing promoter fragments using the JetPrime transfection method according to the manufacturer’s protocol (Polyplus). Expression was allowed to occur at 37 °C for 48 h, after which cells were lysed and analyzed for luciferase levels and activity (Luciferase Assay System, Promega) using a luminometer (1450 microbeta, PerkinElmer). The luminescence values were subtracted from background readings and then normalized against the positive transfection control [pGL4.74(hRluc/TK) Vector, Promega], which expressed Renilla luciferase.
TNS3 CpG island methylation analysis by pyrosequencing
Clinical samples for this study were obtained after nephrectomy from tumor bearing kidneys in patients with histologically verified RCC, performed at the Department of Urology, Umeå University Hospital, between 1988 and 2003. The study was approved by the Umeå University ethical committee and the institutional review board; and informed consent was obtained from each patient. Normal (disease-free) DNA samples were from cell lines derived from peripheral blood lymphocytes of healthy blood donors (HRC1, The Health Protection Agency). The workflow for pyrosequencing analysis of the DNA samples is provided in Figure S5. Genomic DNA was bisulphite converted according to the manufacturer’s instructions (EZ-96 DNA Methylation-Gold™ Kit, Zymo Research). Specific oligonucleotide primers were designed to indiscriminately amplify the desired TNS3 region from both methylated and unmethylated samples post bisulphite conversion (Table S1). Bisulphite-converted DNA was amplified using nested PCR under touchdown conditions: 95 °C for 2 min, 36 cycles of 92 °C for 1 min, annealing temperature reducing from 60 °C to 48 °C; then 72 °C for 1 min, followed by 14 cycles at 47 °C annealing for 1 min, and a final extension at 72 °C for 15 min. PCR products were purified and visualized using a 3% low melting agarose gel before undergoing processing for pyrosequencing (Biotage PSQ 96MA). These steps involved the affinity purification of one strand of the PCR product, which was biotinylated, to enable it to act as a template for pyrosequencing polymerization using a specific sequencing primer; this covered up to a total of 43 individual CpG dinucleotides (through the 2 identified clusters in 3 sequencing reactions).
Cancer cell DNA demethylation by 5-aza-2′-deoxycytidine and TNS3 expression qRT-PCR
Cells from the human kidney cell lines HK-2 and HEK293 were seeded into a 24-well plate in complete medium and allowed to adhere overnight at 37 °C. Forty μM of 5-aza-2′-deoxycytidine was then added to the cells and incubated for a total period of 96h, with cells being lysed at the end. Total RNA was extracted from the lysed cells (GeneJet, Fermentas) followed by reverse transcription (M-MLV, New England Biolabs). Multiplex real-time quantitative RT-PCR (qRT-PCR) was performed on the samples to analyze simultaneous expression of both the TNS3/TENC1 genes and the endogenous control gene B2M. A primer/probe-based system was used for real time detection of PCR amplification (Taqman, Applied Biosystems). For each sample, 1 μl cDNA was mixed with nuclease-free water, qPCR mastermix (Maxima, Fermentas), the primers and probes for TNS3/TENC1 and B2M genes labeled with FAM and VIC fluorophores, respectively, in a total volume of 20 μl. The qPCR was performed under the following cycling conditions: 50 °C for 2 mins, 95 °C for 10 mins and 40 cycles of 95 °C for 15 sec and 60 °C for 60 sec (ABI PRISM 7900HT system, Applied Biosystems). The relative standard curve method was used for quantitative determination of amount of TNS3/TENC1 mRNA generated, using serial dilutions of cDNA from the SNB-19 cell line, which naturally express both Tensin3 and Tensin2, to generate a standard curve. Each measurement was performed in duplicate; individual Ct values were converted into ng RNA from linear regression analysis of the standard curve (Microsoft Excel). Each determined RNA value was then normalized against the endogenous control gene level within the same sample, and the mean of the duplicate values was then calculated.
Statistical analyses
All data were compared using InStat (GraphPad) or SPSS (IBM). Mean differences for functional promoter studies and 5-aza studies were compared using Student’s t-test. Patient samples were compared within the samples for individual differences using a Kruskal–Wallis with a post hoc Dunns test. The individual CpG that were significant were then compared further against the significant other CpGs using a Mann–Whitney U test. Correlations for CpG 2 and CpG 8 against Tensin3 mRNA levels were performed using the Spearman’s Rho correlation test.
| Abbreviations: | ||
| RCC | = | renal cell carcinoma |
| CTCF | = | CCCTC-binding motif |
| YY1 | = | Ying Yang 1 |
| DNMT1 | = | DNA methyltransferase 1 |
| 5-aza | = | 5-aza-2′-deoxycytidine |
Additional material
Download Zip (195.6 KB)Acknowledgments
This work was supported by a research grant from the Swedish Research Council (Vetenskapsrådet; grant no. 70286301) to S.H. and a PhD studentship to J.A.C. from the Institute of Biomedical and Biomolecular Science (IBBS), University of Portsmouth. The authors thank Dr. Derek Brazil (Queen’s University Belfast) for providing the HK-2 cell line.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Supplemental Material
Supplemental materials may be found here: http://www.landesbioscience.com/journals/epigenetics/article/25075
References
- Lo SH. Tensin. Int J Biochem Cell Biol 2004; 36:31 - 4; http://dx.doi.org/10.1016/S1357-2725(03)00171-7; PMID: 14592531
- Hafizi S, Ibraimi F, Dahlbäck B. C1-TEN is a negative regulator of the Akt/PKB signal transduction pathway and inhibits cell survival, proliferation, and migration. FASEB J 2005; 19:971 - 3; PMID: 15817639
- Chuang JZ, Lin DC, Lin S. Molecular cloning, expression, and mapping of the high affinity actin-capping domain of chicken cardiac tensin. J Cell Biol 1995; 128:1095 - 109; http://dx.doi.org/10.1083/jcb.128.6.1095; PMID: 7896874
- Lo SH, Janmey PA, Hartwig JH, Chen LB. Interactions of tensin with actin and identification of its three distinct actin-binding domains. J Cell Biol 1994; 125:1067 - 75; http://dx.doi.org/10.1083/jcb.125.5.1067; PMID: 8195290
- Calderwood DA, Fujioka Y, de Pereda JM, García-Alvarez B, Nakamoto T, Margolis B, et al. Integrin beta cytoplasmic domain interactions with phosphotyrosine-binding domains: a structural prototype for diversity in integrin signaling. Proc Natl Acad Sci U S A 2003; 100:2272 - 7; http://dx.doi.org/10.1073/pnas.262791999; PMID: 12606711
- Hafizi S, Alindri F, Karlsson R, Dahlbäck B. Interaction of Axl receptor tyrosine kinase with C1-TEN, a novel C1 domain-containing protein with homology to tensin. Biochem Biophys Res Commun 2002; 299:793 - 800; http://dx.doi.org/10.1016/S0006-291X(02)02718-3; PMID: 12470648
- Chan LK, Ko FC, Ng IO, Yam JW. Deleted in liver cancer 1 (DLC1) utilizes a novel binding site for Tensin2 PTB domain interaction and is required for tumor-suppressive function. PLoS One 2009; 4:e5572; http://dx.doi.org/10.1371/journal.pone.0005572; PMID: 19440389
- Katz M, Amit I, Citri A, Shay T, Carvalho S, Lavi S, et al. A reciprocal tensin-3-cten switch mediates EGF-driven mammary cell migration. Nat Cell Biol 2007; 9:961 - 9; http://dx.doi.org/10.1038/ncb1622; PMID: 17643115
- Hafizi S, Sernstad E, Swinny JD, Gomez MF, Dahlbäck B. Individual domains of Tensin2 exhibit distinct subcellular localisations and migratory effects. Int J Biochem Cell Biol 2010; 42:52 - 61; http://dx.doi.org/10.1016/j.biocel.2009.09.005; PMID: 19747564
- Martuszewska D, Ljungberg B, Johansson M, Landberg G, Oslakovic C, Dahlbäck B, et al. Tensin3 is a negative regulator of cell migration and all four Tensin family members are downregulated in human kidney cancer. PLoS One 2009; 4:e4350; http://dx.doi.org/10.1371/journal.pone.0004350; PMID: 19194507
- Nishino T, Sasaki N, Chihara M, Nagasaki K, Torigoe D, Kon Y, et al. Distinct distribution of the tensin family in the mouse kidney and small intestine. Exp Anim 2012; 61:525 - 32; http://dx.doi.org/10.1538/expanim.61.525; PMID: 23095816
- Lo SH, Yu QC, Degenstein L, Chen LB, Fuchs E. Progressive kidney degeneration in mice lacking tensin. J Cell Biol 1997; 136:1349 - 61; http://dx.doi.org/10.1083/jcb.136.6.1349; PMID: 9087448
- Chiang MK, Liao YC, Kuwabara Y, Lo SH. Inactivation of tensin3 in mice results in growth retardation and postnatal lethality. Dev Biol 2005; 279:368 - 77; http://dx.doi.org/10.1016/j.ydbio.2004.12.027; PMID: 15733665
- The American Cancer Society. What are the key statistics about kidney cancer. In: The American Cancer Society, ed. Kidney Cancer (Adult) - Renal Cell Carcinoma. Atlanta, GA: The American Cancer Society, 2012.
- Schmidt L, Junker K, Nakaigawa N, Kinjerski T, Weirich G, Miller M, et al. Novel mutations of the MET proto-oncogene in papillary renal carcinomas. Oncogene 1999; 18:2343 - 50; http://dx.doi.org/10.1038/sj.onc.1202547; PMID: 10327054
- Morris MR, Maher ER. Epigenetics of renal cell carcinoma: the path towards new diagnostics and therapeutics. Genome Med 2010; 2:59; http://dx.doi.org/10.1186/gm180; PMID: 20815920
- Larsen F, Gundersen G, Lopez R, Prydz H. CpG islands as gene markers in the human genome. Genomics 1992; 13:1095 - 107; http://dx.doi.org/10.1016/0888-7543(92)90024-M; PMID: 1505946
- Saxonov S, Berg P, Brutlag DL. A genome-wide analysis of CpG dinucleotides in the human genome distinguishes two distinct classes of promoters. Proc Natl Acad Sci U S A 2006; 103:1412 - 7; http://dx.doi.org/10.1073/pnas.0510310103; PMID: 16432200
- Christoph F, Weikert S, Kempkensteffen C, Krause H, Schostak M, Köllermann J, et al. Promoter hypermethylation profile of kidney cancer with new proapoptotic p53 target genes and clinical implications. Clin Cancer Res 2006; 12:5040 - 6; http://dx.doi.org/10.1158/1078-0432.CCR-06-0144; PMID: 16951219
- Esteller M, Fraga MF, Guo M, Garcia-Foncillas J, Hedenfalk I, Godwin AK, et al. DNA methylation patterns in hereditary human cancers mimic sporadic tumorigenesis. Hum Mol Genet 2001; 10:3001 - 7; http://dx.doi.org/10.1093/hmg/10.26.3001; PMID: 11751682
- Moore LE, Nickerson ML, Brennan P, Toro JR, Jaeger E, Rinsky J, et al. Von Hippel-Lindau (VHL) inactivation in sporadic clear cell renal cancer: associations with germline VHL polymorphisms and etiologic risk factors. PLoS Genet 2011; 7:e1002312; http://dx.doi.org/10.1371/journal.pgen.1002312; PMID: 22022277
- Ibanez de Caceres I, Dulaimi E, Hoffman AM, Al-Saleem T, Uzzo RG, Cairns P. Identification of novel target genes by an epigenetic reactivation screen of renal cancer. Cancer Res 2006; 66:5021 - 8; http://dx.doi.org/10.1158/0008-5472.CAN-05-3365; PMID: 16707423
- Rice P, Longden I, Bleasby A. EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet 2000; 16:276 - 7; http://dx.doi.org/10.1016/S0168-9525(00)02024-2; PMID: 10827456
- Gardiner-Garden M, Frommer M. CpG islands in vertebrate genomes. J Mol Biol 1987; 196:261 - 82; http://dx.doi.org/10.1016/0022-2836(87)90689-9; PMID: 3656447
- Li L-C, Dahiya R. MethPrimer: designing primers for methylation PCRs. Bioinformatics 2002; 18:1427 - 31; http://dx.doi.org/10.1093/bioinformatics/18.11.1427; PMID: 12424112
- Ryschich E, Lizdenis P, Ittrich C, Benner A, Stahl S, Hamann A, et al. Molecular fingerprinting and autocrine growth regulation of endothelial cells in a murine model of hepatocellular carcinoma. Cancer Res 2006; 66:198 - 211; http://dx.doi.org/10.1158/0008-5472.CAN-05-1636; PMID: 16397233
- Santin AD, Zhan F, Cane’ S, Bellone S, Palmieri M, Thomas M, et al. Gene expression fingerprint of uterine serous papillary carcinoma: identification of novel molecular markers for uterine serous cancer diagnosis and therapy. Br J Cancer 2005; 92:1561 - 73; http://dx.doi.org/10.1038/sj.bjc.6602480; PMID: 15785748
- Dahl E, Sadr-Nabavi A, Klopocki E, Betz B, Grube S, Kreutzfeld R, et al. Systematic identification and molecular characterization of genes differentially expressed in breast and ovarian cancer. J Pathol 2005; 205:21 - 8; http://dx.doi.org/10.1002/path.1687; PMID: 15586368
- Kim KE, Song H, Kim TS, Yoon D, Kim CW, Bang SI, et al. Interleukin-18 is a critical factor for vascular endothelial growth factor-enhanced migration in human gastric cancer cell lines. Oncogene 2007; 26:1468 - 76; http://dx.doi.org/10.1038/sj.onc.1209926; PMID: 17001321
- Albasri A, Seth R, Jackson D, Benhasouna A, Crook S, Nateri AS, et al. C-terminal Tensin-like (CTEN) is an oncogene which alters cell motility possibly through repression of E-cadherin in colorectal cancer. J Pathol 2009; 218:57 - 65; http://dx.doi.org/10.1002/path.2508; PMID: 19214987
- Sasaki H, Yukiue H, Kobayashi Y, Fukai I, Fujii Y. Cten mRNA expression is correlated with tumor progression in thymoma. Tumour Biol 2003; 24:271 - 4; http://dx.doi.org/10.1159/000076141; PMID: 15001839
- Lo SH, Lo TB. Cten, a COOH-terminal tensin-like protein with prostate restricted expression, is down-regulated in prostate cancer. Cancer Res 2002; 62:4217 - 21; PMID: 12154022
- Albasri A, Aleskandarany M, Benhasouna A, Powe DG, Ellis IO, Ilyas M, et al. CTEN (C-terminal tensin-like), a novel oncogene overexpressed in invasive breast carcinoma of poor prognosis. Breast Cancer Res Treat 2011; 126:47 - 54; http://dx.doi.org/10.1007/s10549-010-0890-3; PMID: 20390342
- Li Y, Mizokami A, Izumi K, Narimoto K, Shima T, Zhang J, et al. CTEN/tensin 4 expression induces sensitivity to paclitaxel in prostate cancer. Prostate 2010; 70:48 - 60; http://dx.doi.org/10.1002/pros.21037; PMID: 19725034
- Liao YC, Chen NT, Shih YP, Dong Y, Lo SH. Up-regulation of C-terminal tensin-like molecule promotes the tumorigenicity of colon cancer through beta-catenin. Cancer Res 2009; 69:4563 - 6; http://dx.doi.org/10.1158/0008-5472.CAN-09-0117; PMID: 19487278
- Sasaki H, Moriyama S, Mizuno K, Yukiue H, Konishi A, Yano M, et al. Cten mRNA expression was correlated with tumor progression in lung cancers. Lung Cancer 2003; 40:151 - 5; http://dx.doi.org/10.1016/S0169-5002(03)00037-0; PMID: 12711115
- The EPC, ENCODE Project Consortium. A user’s guide to the encyclopedia of DNA elements (ENCODE). PLoS Biol 2011; 9:e1001046; http://dx.doi.org/10.1371/journal.pbio.1001046; PMID: 21526222
- Biel M, Wascholowski V, Giannis A. Epigenetics--an epicenter of gene regulation: histones and histone-modifying enzymes. Angew Chem Int Ed Engl 2005; 44:3186 - 216; http://dx.doi.org/10.1002/anie.200461346; PMID: 15898057
- Li B, Carey M, Workman JL. The role of chromatin during transcription. Cell 2007; 128:707 - 19; http://dx.doi.org/10.1016/j.cell.2007.01.015; PMID: 17320508
- Kim TH, Abdullaev ZK, Smith AD, Ching KA, Loukinov DI, Green RD, et al. Analysis of the vertebrate insulator protein CTCF-binding sites in the human genome. Cell 2007; 128:1231 - 45; http://dx.doi.org/10.1016/j.cell.2006.12.048; PMID: 17382889
- Yang J, Corces VG. Insulators, long-range interactions, and genome function. Curr Opin Genet Dev 2012; 22:86 - 92; http://dx.doi.org/10.1016/j.gde.2011.12.007; PMID: 22265227
- Bell LA, Ryan KM. Life and death decisions by E2F-1. Cell Death Differ 2004; 11:137 - 42; http://dx.doi.org/10.1038/sj.cdd.4401324; PMID: 14526389
- Zhang Q, Stovall DB, Inoue K, Sui G. The oncogenic role of Yin Yang 1. Crit Rev Oncog 2011; 16:163 - 97; http://dx.doi.org/10.1615/CritRevOncog.v16.i3-4.30; PMID: 22248053
- Abdulrazzak H, Noro N, Simons JP, Goldspink G, Barnard EA, Górecki DC. Structural diversity despite strong evolutionary conservation in the 5′-untranslated region of the P-type dystrophin transcript. Mol Cell Neurosci 2001; 17:500 - 13; http://dx.doi.org/10.1006/mcne.2000.0950; PMID: 11273645
- Christman JK. 5-Azacytidine and 5-aza-2′-deoxycytidine as inhibitors of DNA methylation: mechanistic studies and their implications for cancer therapy. Oncogene 2002; 21:5483 - 95; http://dx.doi.org/10.1038/sj.onc.1205699; PMID: 12154409
- Ryan MJ, Johnson G, Kirk J, Fuerstenberg SM, Zager RA, Torok-Storb B. HK-2: an immortalized proximal tubule epithelial cell line from normal adult human kidney. Kidney Int 1994; 45:48 - 57; http://dx.doi.org/10.1038/ki.1994.6; PMID: 8127021
- Maherally Z, Smith JR, An Q, Pilkington GJ. Receptors for hyaluronic acid and poliovirus: a combinatorial role in glioma invasion?. PLoS One 2012; 7:e30691; http://dx.doi.org/10.1371/journal.pone.0030691; PMID: 22363471