Abstract
Previous studies have uncovered several transcription factors that determine biological alterations in tumor cells to execute the invasion-metastasis cascade, including the epithelial-mesenchymal transition (EMT). We sought to investigate the role of miR-21 in colorectal cancer regulation. For this purpose, miR-21 expression was quantified in a panel of colorectal cancer cell lines and clinical specimens. High expression was found in cell lines with EMT properties and in the vast majority of human tumor specimens. We demonstrate in a cell-specific manner the occupancy of MIR-21 gene promoter by AP-1 and ETS1 transcription factors and, for the first time, the pattern of histone posttranslational modifications necessary for miR-21 overexpression. We also show that Integrin-β4 (ITGβ4), exclusively expressed in polarized epithelial cells, is a novel miR-21 target gene and plays a role in the regulation of EMT, since it is remarkably de-repressed after transient miR-21 silencing and downregulated after miR-21 overexpression. miR-21-dependent change of ITGβ4 expression significantly affects cell migration properties of colon cancer cells. Finally, in a subgroup of tumor specimens, ROC curve analysis performed on quantitative PCR data sets for miR-21, ITGβ4, and PDCD4 shows that the combination of high miR-21 with low ITGβ4 and PDCD4 expression is able to predict presence of metastasis. In conclusion, miR-21 is a key player in oncogenic EMT, its overexpression is controlled by the cooperation of genetic and epigenetic alterations, and its levels, along with ITGβ4 and PDCD4 expression, could be exploited as a prognostic tool for CRC metastasis.
Introduction
Features of malignant tumors include invasion and metastasis. Metastasis constitutes a complex process during which primary tumor cells invade adjacent normal tissues and metastasize to distant organs, where they give birth to secondary tumors.Citation1 Loss of E-cadherin expression is considered crucial in the transition of non-invasive adenoma to invasive carcinoma and represents a fundamental step of epithelial-mesenchymal transition (EMT).Citation2,Citation3 In recent years, enormous efforts have been done to elucidate the mechanisms of the metastatic process, as well as to identify which genes are involved. Moreover, it has become evident that abnormalities in non-coding genes, including microRNAs (miRNAs), can contribute to cancer pathogenesis. Particularly, the effect of miRNAs on cell invasion and migration has been documented,Citation4 even though the mechanisms involved and the functionality of miRNAs in cancer are largely unknown.Citation5 Studies have uncovered several transcription factors that can affect or even determine many biological changes needed to execute the initial steps of the invasion-metastasis cascade.Citation6-Citation8 For example, the transcription factor AP-1, a heterodimeric complex mainly composed of JUN and FOS families, is a regulator of several cancer hallmarks.Citation9–Citation11 Expression of AP-1 components, FRA1 and JUN, is induced by the RAF/MEK/ERK mitogenic cascade,Citation12 a distinct signaling pathway triggered by the binding of RAS effector proteins to RAF kinase and/or phosphatidylinositol 3-kinase (PI3K).Citation13
miR-21 is considered an oncomir; indeed, unlike other miRNAs, it is frequently upregulated in the majority of solid tumors,Citation14 including breast,Citation15 prostate,Citation16 colon,Citation17 and lung cancer,Citation18 as well as glioblastoma.Citation19 More importantly, miR-21 expression level is associated with established prognostic factors and constitutes a significant predictor of survival in colorectal cancer (CRC)Citation20,Citation21 and in non-small-cell lung cancer.Citation18 Evidence indicates that deregulation of the RAS pathway,Citation22,Citation23 epigenetic alterations,Citation24,Citation25 mutations,Citation26 DNA copy number abnormalities,Citation27 and defects in the miRNA biogenesis machineryCitation28 might contribute to deregulation of miRNAs levels in human cancer. Moreover, it has recently been proven that miRNAs, and in particular miR-21, play a role in regulatory circuits involving epigenetic switches, required for the transformation of cancer cells.Citation29 Interestingly, these circuits can be transiently activated by specific stimuli and propagated for many generations in the absence of the initiating signal.Citation30
Transcription activation of gene promoters largely depends on chromatin conformation.Citation31 In fact, euchromatin is associated with distinct active domains, whereas transcriptionally inactive domains lie within the heterochromatin. Regulation of chromatin status is achieved by epigenetic mechanisms, such as posttranslational modifications of histone tails operated by modifier enzymes on specific amino acid residues. Important histone modifications include, among others, acetylation, methylation, and phosphorylation. Deregulation of histone modifications and the respective modifier enzymes have been associated with cancer,Citation32 although no studies have focused and investigated the aberrant histone modifications that may be responsible for miR-21 upregulation in CRC.
Epithelial tissues, such as colon mucosa, are characterized by a well-defined tissue architecture that largely depends on the expression of specific cell surface adhesion molecules (such as integrins, cadherins, and others), that interact with extracellular matrix and neighboring cells. During malignant progression, the expression and sub-cellular localization of several cell adhesion molecules change as a consequence of EMT. Integrins are transmembrane αβ-heterodimer glycoproteins, constituting a major class of receptors that can interact with extracellular matrix molecules and mediate cell-matrix interactions. Many integrins are expressed by human intestinal epithelial cells.Citation33,Citation34 Integrin β4 (ITGβ4) forms, together with ITGα6, a single functional heterodimer, named ITGα6β4. Studies have shown that distinct forms of the α6 and β4 subunits are expressed in the intestine tissue and, more importantly, various forms of α6β4 can differentially regulate intestinal epithelial cell functions under both normal and pathological conditions.Citation35 In CRC, expression of ITGβ4 subunit has been characterized in a number of studies.Citation36-Citation38 Although these studies report uniform expression of ITGβ4 at the base of epithelial cells in the normal colon mucosa, the analysis of its expression in malignancies has led to conflicting results. Indeed, ITGβ4 expression was found to be reduced or even lost,Citation36 maintained,Citation38 or heterogeneously increasedCitation37 in carcinomas. Interestingly, Sordat et al. described a particular expression pattern for ITGβ4, which was maintained in well-differentiated carcinomas and decreased in moderately and poorly differentiated carcinomas.Citation38 The last findings indicate that ITGβ4 is critical for a correct and functional tissue architecture, since tumor tissues where its expression is low or lost present the most aggressive phenotype.
In order to better understand the miR-21 deregulation and its implication in EMT, we focused on how transcriptional and epigenetic mechanisms might cooperate to deregulate miR-21 in CRC. For this purpose, miR-21 was quantified in a panel of CRC cell lines and in stably transfected Caco-2 clones bearing a mutated Harvey-RAS (HRAS) oncogene Caco-H2.Citation39 Chromatin conformation status on MIR21 gene promoter was investigated, as well as the occupancy of the MIR21 gene promoter by AP-1 and ETS1 transcription factors. Using anti-miR-21 and mimic-miR-21 we investigated the effect of miR-21 inhibition and overexpression on cell-migration and -viability and sought for new target genes. ITGβ4 protein expression was affected positively by miR-21 silencing and negatively by miR-21 overexpression and is proposed as a new miR-21 target. Finally, the prognostic performance of a combined 3-gene assay (MIR21, ITGβ4, and PDCD4) was demonstrated to be effective on a subgroup of CRC patients toward prediction of metastatic disease.
Results
miR-21 is overexpressed in CRC cell lines with high metastatic potential and EMT traits
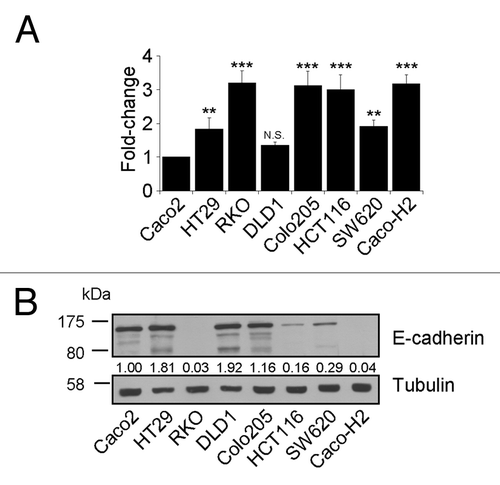
We quantified miR-21 levels in relation to the snRNA RNU6B in CRC cell lines with distinct properties, as well as in Caco-H2 (). Caco-2 and DLD-1 cells were shown to express low levels of miR-21 in comparison with all other cell lines, with Caco-2 intermediate adenoma cell line showing the lowest. For this reason, relative miR-21 levels in all cell lines were normalized against miR-21 levels in Caco-2 cells. On the other hand, levels of miR-21 in HT-29 and SW620 colon cancer cells were 1.5-fold higher, compared with miR-21 levels in Caco-2 cells. Moreover, HCT116, Colo-205, Caco-H2, and RKO cells presented a very high expression of miR-21, about 3-fold compared with the calibrator cell line (Caco-2). In EMT cell lines showing very low levels of the epithelial marker E-cadherin (; Table S1), high miR-21 expression was detected, with the exception of Colo-205 cell line.
Figure 1. Expression levels of mature miR-21 and the epithelial marker E-Cadherin in CRC cell lines. (A) Quantitative PCR analysis showed high levels of mature miR-21 in cell lines with mesenchymal traits. (B) E-Cadherin protein levels are particularly low in cells overexpressing miR-21. Stars on each bar indicate the P value (t test) calculated with respect to the expression value of Caco-2 cell line. Significant differences: ***P ≤ 0,001; **P ≤ 0,005; N.S. not significant.
AP-1 and ETS1 bind the promoter of the MIR21 gene with different affinity in colorectal cancer cell lines with distinct EMT properties
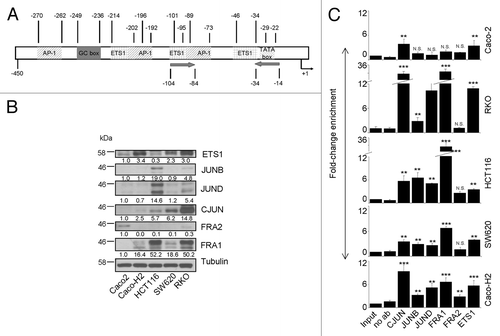
Loss of E-cadherin is recognized as a marker of EMTCitation2,Citation3 and is associated with gain of cell migration ability. Figure S1 shows the morphology of Caco-H2, SW620, HCT116, RKO, and the control Caco-2 cell line. While Caco-2 cells form small cell aggregates and do not migrate (Table S1), the other examined cell lines have the tendency to grow singularly with plasma membrane protrusion and express a mesenchymal marker such as Vimentin. Furthermore, compared with Caco-2 they have superior migration ability. All together these characteristics represent clear traits of EMT phenotype. A previous studyCitation40 demonstrated that miR-21 overexpression is linked to invasion and metastasis, but little is known about which factors and pathways are involved in miR-21 upregulation. Thus, we sought to identify transcription factors that might account for miR-21 overexpression, with consequent enhancement of cell mobility, using cell models with EMT properties and migration ability. Toward this direction, we determined the occupancy of the MIR21 gene promoter by AP-1 and ETS1 in the above mentioned EMT CRC cell lines. shows a schematic representation of the MIR21 gene promoter based on the structure that Fujita et al. proposed,Citation41 possessing three putative AP-1-binding sites and two potential ETS1-binding sites. First, we examined the protein levels of AP-1 family (CJUN, JUNB, JUND, FRA1, and FRA2) and ETS1 transcription factors (). All of them, except for FRA2, were expressed almost exclusively in cell lines with an invasive phenotype. Therefore, we performed a detailed ChIP assay to study the binding properties of each transcription factor on the miR-21 promoter (). This analysis revealed that both AP-1 and ETS1 transcription factors bind the miR-21 promoter in CRC cell lines, though with different affinity. In particular, FRA1 shows very strong affinity for miR-21 gene promoter, mainly in HCT116 and RKO cells, as ChIP unveils a 29- and 26-fold enrichment (), respectively, as compared with control-input DNA. CJUN is also strongly bound to the miR-21 promoter in RKO cells, presenting a 26-fold enrichment. The other members of the JUN family overall present a significant binding on miR-21 promoter in EMT cell lines, compared with Caco-2 control cells. Similar was the occupancy for ETS1; in Caco-2 cells the enrichment is ~3-fold as compared with input DNA, whereas in EMT cells it ranges from more that 4- in HCT116 cells up to 8-fold in RKO cells, as compared with input DNA. Moreover, to provide further evidence that pathways regulating AP-1/ETS factors can also affect miR-21 expression, the MAPK pathway was blocked in HCT116 cells by a treatment with the MEK inhibitor UO126 for a time course of 24 h. A considerable downregulation in pri-miR-21 precursor expression was detected within the first few hours after treatment, coinciding with the reduction of ERK phosphorylation (Fig. S2). These data point to a transcriptional effect of the MAPK pathway on miR-21 expression. No reduction in the biogenesis of mature miR-21 was detected, probably due to the high accumulation of mature products in the cell body and/or increased stability of the mature species in this particular cell line.
Figure 2. Expression and occupancy of MIR-21 gene promoter by AP-1 and ETS1 transcription factors and its regulation by MAPK pathway in CRC cell lines with EMT properties. (A) Schematic representation of the MIR-21 gene promoter. AP-1, ETS1, and others regulatory elements are shown. Numbers represent the position of regulatory sequences with respect to transcription start site. Arrows indicate the position of forward and reverse primers used for ChIP experiments. (B) Protein expression of ETS1 and major AP-1 family components in selected cell lines with EMT phenotype. (C) ChIP-coupled quantitative PCR analysis of enrichment of transcription factors at MIR-21 promoter. FRA1 and CJUN showed strong binding in all EMT cell lines compared with Caco-2 cells. Enrichment for each transcription factor was normalized to input DNA using the ΔΔCt method. Stars on each bar indicate the p value (t-test) calculated with respect to Input DNA. Significant differences: ***P ≤ 0,001; **P ≤ 0,005; N.S., not significant
AP-1 and ETS1 transcription factors regulate transcription of the MIR21 gene
We studied the transcriptional regulation potential of miR-21 by AP-1 and ETS1 siRNA in SW620, HCT116, and Caco-H2 cell lines (Fig. S3A). CJUN/FRA1/JUNB and ETS1 silencing reduced miR-21 gene expression with different efficiency, both at primary transcript and at mature miR-21 level in the studied cell lines (). pri-miR-21 levels showed a covariance with mature miR-21 levels, both showing remarkable decrease in most cases in almost all cell line systems. In HCT116 cells we verified by ChIP analysis the displacement from miR-21 promoter of CJUN and FRA1 after siRNA. JUN enrichment decreased from 8.5 to 5.2, whereas for FRA1 the reduction was more consistent from 29.66 to 7.2 (Fig. S3B). These results showed that miR-21 transcription was reduced as a direct consequence of CJUN/FRA1 lowering from miR-21 promoter.
Figure 3. Regulation of MIR-21 gene expression by selected AP-1 and ETS1 transcription factors. (A) Quantitative PCR of pri-miR-21 and mature miR-21 in three EMT cell lines transiently transfected with siRNA against CJUN, JUNB, FRA1, and ETS1 genes. Both species of miR-21 gene were reduced after transcription factor silencing. (B) Quantitative PCR of pri-miR-21 and mature miR-21 in Caco-2 cells transfected with plasmid vectors carrying ETS1 and CJUN cDNA. Upon ectopic overexpression of ETS1 and CJUN proteins, levels of pri-miR-21 and mature miR-21 were increased. Stars on each bar indicate the p value (t-test) calculated with respect to untreated samples. Significant differences: ***P ≤ 0,001; **P ≤ 0,005; N.S., not significant
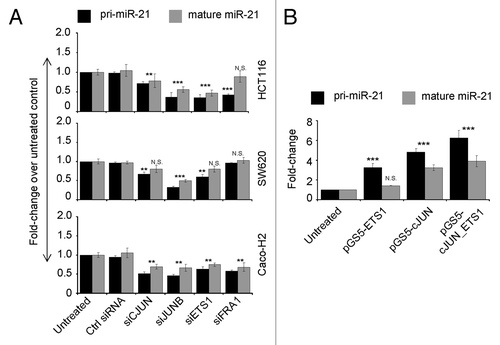
We next performed overexpression of CJUN and ETS1, each one alone or in combination, in Caco-2 cells (Fig. S4). This overexpression resulted in a remarkable increase of pri-miR-21 levels ranging from 3 to 6 fold, which was higher after CJUN overexpression and even higher after co-overexpression of CJUN and ETS1 proteins (). On the other hand, as for siRNA experiments, levels of the mature miR-21 did not increase as much as pri-miR-21 levels. However, after ETS1 overexpression, yet a 1.4-fold increase in mature miR-21 levels was observed in Caco-2 cells, as well as an almost 3.5-fold increase in miR-21 levels after CJUN overexpression. In CJUN and ETS1 co-transfection, overexpression of both transcription factors resulted in a considerable near to 4-fold induction of the mature form of miR-21 ().
High levels of activating histone posttranslational modification on the MIR21 gene promoter correlate with miR-21 expression
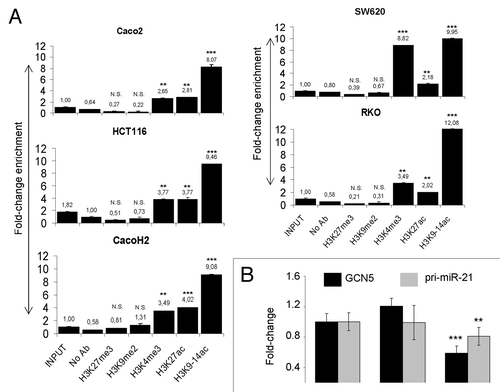
Chromatin conformation status plays a major role in gene expression.Citation31 To verify if across the cell lines analyzed the different transcription factor binding affinity to miR-21 promoter is dependent on epigenetic mechanisms, we performed ChIP analysis using antibodies for histone modifications associated with active transcription state (H3K9-14ac, H3K3me3, and H3K27ac) and with inactive transcription state (H3K27me3 and H3K9me2). We found complete absence of inactive marks such as H3K27me3 and H3K9me2 on miR-21 promoter in all cell lines analyzed (). On the other hand, histone modifications such as H3K9-14ac, H3K4me3, and H3K27ac, associated with active transcription, were higher on miR-21 promoter in EMT cells compared with control Caco-2 cells. Correlation between transcription factor binding and histone modification revealed that high histone acetylation is associated with AP-1 and ETS1 binding on the miR-21 promoter. The relationship between miR-21 promoter acetylation and miR-21 expression was studied using siRNA vs. the major histone acetyl-transferase enzyme GCN5. For the selection of a cell system to use in this experiment we first performed a protein expression analysis of GCN5 in all cell lines by WB. GCN5 protein level was found particularly high in RKO and SW620 cell lines (Fig. S5A). Silencing of GCN5 protein in RKO cells resulted in a reduction of pri-miR-21 mRNA ().
Figure 4. Histone posttranslational modifications on MIR-21 gene promoter and their effects on miR-21 expression. (A) ChIP-coupled quantitative PCR analysis related to the enrichment of activating (H3K9–14ac, H3K27ac, and H3K4me3) and repressing (H3K27me3, H3K9me2) histone marks on MIR-21 promoter. (B) Quantitative PCR analysis of pri-miR-21 abundance in RKO cells treated with siRNA vs. the histone acetyltransferase enzyme GCN5. RNA was extracted 48 h after siGCN5 transfection. Stars on each bar indicate the p value (t-test) calculated with respect to input DNA or untreated samples. Significant differences: ***P ≤ 0,001; **P ≤ 0,005; N.S., not significant. Numbers above the bars represent fold change.
miR-21 inhibition reduces the migrating properties of colon cancer cells with EMT phenotype and de-represses ITGβ4, its new target gene
We blocked the activity of endogenous mature miR-21 using a specific miR-21 inhibitor based on the LNA technology in HCT116 and RKO cell lines. Twenty-four hours after transfection, protein levels of a validated miR-21 target (PTEN) increased, confirming the functional blocking of miR-21 (Fig. S5B). To verify the role of miR-21 in cell migration, HCT116 and RKO cells were transiently transfected with anti-miR-21 and their migration capacity was measured using transwell systems. Inhibition of mature miR-21 caused reduction of migration capacity by roughly 40% in both cell lines (). Furthermore, viability tests (SRB and MTT assays) confirmed that the effect on cell migration was due to the reduction of miR-21 activity and not to cell number reduction. In fact, anti-miR-21 treatment affected cell viability only marginally after 24 h, with a slight effect (~15% less cell) after a 48h anti-miR-21 transfection (Fig. S5C). The role of miR-21 in cell migration was further investigated with a gain of function approach. Because of the inability of Caco-2 to move (Table S1) and to migrate on transwell system, the link among miR-21 overexpression and migration was analyzed using the DLD1 cell line, that presents miR-21 and E-cadherin steady-state expression levels similar to Caco-2, but DLD1 cells are able to migrate (). In DLD1 cells, miR-21 was overexpressed using mirVana mimic technology. Twenty-four hours after transfection miR-21 was upregulated by 16.45-fold (Fig. S6A) and migration and wound healing assays were set allowing cells to migrate for additionally 24 h. Overexpression of miR-21 caused an increase of DLD1 cell migration on transwell system by 35% (). Gain of DLD1 cell mobility and cell-phenotype changes (elongated cell-body typical of mesenchymal cells) were also observed on wound healing assay (Fig. S6B).
Figure 5. miR-21 posttranscriptionally regulates ITGβ4 and controls cell migration. (A) HCT116 and RKO were transfected with miR-21 inhibitor and their migratory properties were tested with transwell systems. Reduction of migratory ability was observed and quantified in anti-miR-21 treated cells (bottom histograms). Quantification of migratory ability was done by counting the number of cells that passed through the polycarbonate membrane of transwells in three randomly chosen areas for each experimental point considering parental cells as control (bottom histograms). (B) Overexpression of mature miR-21 in DLD1 using mirVana mimic-miR-21 enhanced cell migration. (C) ITGβ4 mRNA and (D) ITGβ4 protein expression increased in HCT116 and RKO transfected with miR-21 inhibitor. (E) Confocal microscopy pictures of HCT116 cells treated with anti-miR-21. Nuclei have been counterstained with Hoechst (blue). ITGβ4 (red), phase contrast images and nuclei staining Hoechst are also showed in a unique stack. Band 20 μm. (F) Overexpression of mature miR-21 in Caco-2 using mirVana mimic-miR-21 caused ITGβ4 protein downregulation. (G) ITGβ4 protein and mRNA levels in CRC cell lines. Stars on each bar indicate the P value (t-test) calculated with respect to parental, untreated samples or over ITGβ4 mRNA level of Caco-2 cell lines. Significant differences: ***P ≤ 0,001; **P ≤ 0,005; N.S. not significant
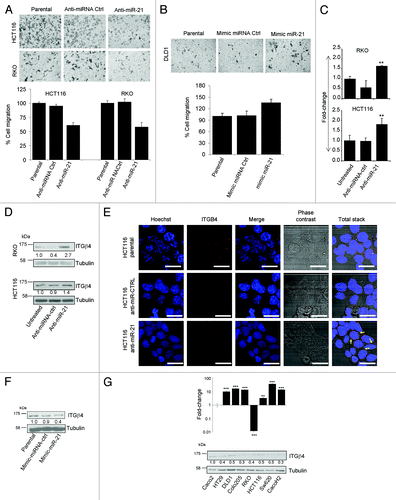
Expression analysis (data not shown) on a panel of genes related to cell migration was performed in HCT116 and RKO cells transiently transfected with anti-miR-21 in order to identify potential target genes that are regulated by miR-21. ITGβ4 mRNA was found to be enhanced by nearly 2-fold in cells treated with anti-miR-21 (). De-repression of ITGβ4 was confirmed by WB analysis in cell lines with EMT properties (). Furthermore, a confocal microscopy scan of HCT116 cells treated with anti-miR-21 showed a clearly de-repression of ITGβ4 (dots, ) and most importantly provided a rational explanation for cell migration inhibition. Indeed, in anti-miR-21-treated HCT116 cells the sub-cellular localization of several dots appeared at the cell junctions (arrows ), resulting in a more tight cell-cell contact that impairs cell-mobility. To corroborate this finding miR-21 was overexpressed in Caco-2 control cell line and a reduction of 60% of ITGβ4 protein was detected 24 h post transfection ().
The entire coding sequence of ITGβ4 mRNA was scanned for the presence of miR-21 putative target sites using RegRNA software.Citation42 Several putative miR-21 binding sites were identified; those with high score are reported in Figure S7. Interestingly, not only the major miR-21 strand (hsa-miR-21-5p) is able to target ITGβ4 coding sequence at the position 735–757, but also the minor strand named hsa-miR21–3p (also known as hsa-miR-21*). Furthermore, evaluation of ITGβ4 mRNA and protein levels on 8 CRC cell lines confirmed that ITGβ4 is posttranscriptionally regulated in colon cancer. In fact, although with respect to Caco-2 cell line all other cell lines, except RKO, express high level of ITGβ4 mRNA (>10-fold), ITGβ4 protein levels were lower in comparison with Caco-2 ().
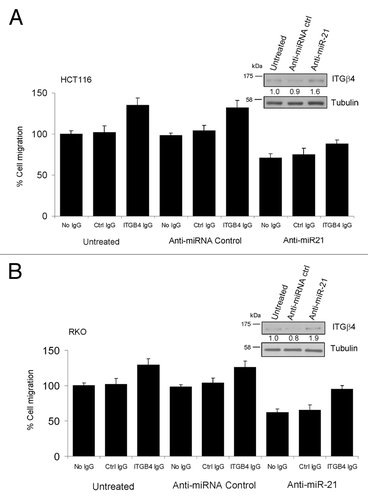
Finally, to prove that the axis miR-21-ITGβ4 controls, at least in part, cell migration in CRC cell lines, we used two different approaches. RKO and HCT116 were: (1) co-transfected with anti-miR-21 and siITGβ4, seeded on transwell and cell migration was examined; (2) transfected with anti-miR-21, seeded on transwell in presence of anti-ITGβ4 blocking antibody and cell migration was examined. shows results of the last approach described above in RKO and HCT116 cell lines. Interestingly, ITGβ4 silencing (data not shown) and functional blocking () separately were able to enhance cell mobility in control cells and most importantly restore migration by ~50% in anti-miR-21-treated RKO cells (). These two assays, together with the miR-21 overexpression in Caco-2 and DLD1 (), demonstrate that ITGB4 is a target of miR-21 and both play a role in tumor cell migration.
Figure 6. Functional inhibition of ITGβ4 protein in CRC cell lines reverts anti-miR-21 effects on cancer cell migration. (A and B) HCT116 and RKO cell lines were transfected with anti-miR-21 and seeded on transwell together with 50 ng/ml of anti-ITGβ4 blocking antibody and migration ability was evaluated as described above. In both cases, inhibition of ITGβ4 protein function resulted in cell migration enhancement. Levels of ITGβ4 protein are shown for each experiment.
miR-21, ITGΒ4 and PDCD4 expression levels predict presence of metastasis in CRC specimens
A quantitative PCR analysis was performed on 64 specimens of colon adenocarcinoma tumors and 9 normal colon mucosa controls. Overall, miR-21 was upregulated in 82.8% (53 out of 64) of tumors compared with normal tissues (P = 0.0001) with a fold-change median >3 (; ). ITGβ4 expression was variable. In tumor tissues, variance ranged from 0.22- to 160-fold, with a median close to the normal tissues value, 1.96 and 1.35, respectively (P = 0.3829) (; ). Programmed cell death 4 (PDCD4) is a transformation suppressor that inhibits tumor neoplastic transformation and is involved in AP-1-dependent transcription required for transformation. Studies have shown that, in CRC, PDCD4 protein level is controlled by miR-21, but PDCD4 mRNA degradation by miR-21 is not clear.Citation40,Citation43,Citation44 To further validate the miR-21-related PDCD4 regulation in tumor tissues, we examined its levels in the group of specimens analyzed for miR-21 and ITGβ4 expression. PDCD4 mRNA levels, unlike ITGβ4, were low or very low in the vast majority of tumor tissues analyzed, median value 0.28 (P < 0.0001) (; ) in contrast with a previous study.Citation40 We evaluated, using receiver operating characteristic (ROC) curve and quantitative PCR data sets, the usefulness of miR-21- and target-expression as prognostic tools for metastatic disease prediction. ROC curves performed using a single-gene test on undivided specimens are shown in . The area under curve (AUC) for each gene alone was close to 0.5, discriminating power was not better than by chance (table in ). Quantitative data were combined using logistic regression and the performance of a 3-gene prognostic assay was tested on all samples. Once again, on undivided specimens the prognostic accuracy was poor AUC = 0.55, P = 0.5 (Fig. S8A). We assumed that ITGβ4 might be critical on the initiation of metastatic cascade when downregulated. Therefore, tumor samples were grouped in three cohorts (low [11/64], normal [15/64], and high [38/64]) based on ITGβ4 expression (cut off <0.8-fold) and statistical analysis was performed on each group. The performance of ROC curves, both individually and in combination, was optimal in the low ITGβ4 group (). The best AUC value (>0.86, P < 0.01) for this group was obtained when miR-21, ITGβ4, and PDCD4 genes were combined using logistic regression in a unique 3-gene prognostic assay (). The same analysis performed on cohorts with normal and high ITGβ4 mRNA did not show any significant predictive ability (Fig. S8B and C).
Figure 7. Expression analysis of miR-21, its target genes in human CRC specimens and performance of a 3-gene metastasis prognostic assay. (A) Each panel compares the expression values of miR-21, ITGβ4, and PDCD4 in normal and in CRC tissues using the box-and-whisker plot. (B) Single-gene quantitative PCR data sets were subjected to ROC curve analysis and the performance to discriminate between metastatic and non-metastatic patients was tested. Values of AUC are provided in the enclosed table. (C) ROC curve analysis performed using samples with low level of ITGβ4. AUC values for each gene are provided in the enclosed table. (D) For the group of samples with low ITGβ4 mRNA level, quantitative 3-gene PCR data sets were combined using logistic regression in a unique 3-gene assay and the resulting predicted probabilities were subjected to ROC curve analysis. Sensitivity, specificity, cut-off, P value, standard error (SE), and AUC values are enclosed.
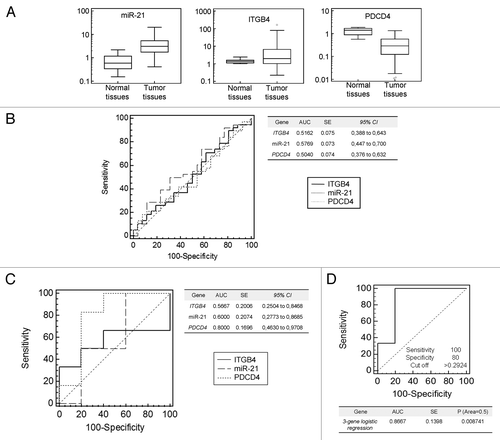
Table 1. Statistical analysis values from quantitative PCR data sets for miR-21, ITGβ4, and PDCD4 in human normal and tumor colon tissues
Discussion
Oncogenic transformation is generally associated with enhancement of endogenous AP-1 activity through various pathways, and this strongly contributes to malignant potential.Citation45 However, not all regulatory networks and target genes depending on AP-1 activity are well investigated. Previous studies have shown that binding of both FOS and JUN to miR-21 promoter region is increased after phorbol 12-myristate 13-acetate PMA stimulation, and that JUNB, JUND, and PU.1 (ETS1 family) also bind MIR21 gene promoter in cancer cells.Citation40 Nevertheless, miR-21 transcription can also be induced by STAT3 due to two binding sites on its gene promoter, or by AKT pathway.Citation29,Citation46 Furthermore, in vitro treatment with the DNA demethylating agent 5-aza-2-deoxycytidineCitation25 and the histone deacetylase inhibitor 4-phenylbutyric acid, which affected miR-21 global expression, suggested an involvement of epigenetic mechanisms in deregulation of miRNAs expression in cancer.Citation47
miR-21 is regulated by onco-epigenetic pathways
We have investigated in detail miR-21 promoter occupancy by AP-1 and ETS transcription factors by performing ChIP, silencing, and overexpressing these transcription factors in colon cancer cell lines with distinct EMT traits. We did not observe, in all cell lines used, concomitant variation between pri-miR-21 and mature miR-21 levels in siRNA or transcription factor overexpression experiments, probably due to the delay between transcription and maturation process and/or the presence of an additional protective feedback mechanism. Nevertheless, CJUN, FRA1, and ETS1 were found binding the MIR21gene promoter. In particular, we confirmed that FRA1Citation22 shows very strong affinity for MIR21 gene promoter in cancer cells with invasive phenotype, such as HCT116 and RKO. These data suggest a possible mechanism through which FRA1 might act in cancer cells, since FRA1 is able to enhance cancer cell invasivenessCitation48 and is upregulated in several human malignances, including breast cancer.Citation49 Another important novel finding of our work is that FRA1 and other AP-1 components together with the ETS1 transcription factor increase their occupancy on the MIR21 gene promoter in Caco-H2, compared with the parental Caco-2 cells. Although a potential synergy between members of the two families of transcription factors was suggested by two independent studies,Citation22,Citation45 this study provides proof that RAS pathway controls miR-21 expression via AP-1 and ETS1 transcription factors that are overexpressed in a cell line (Caco-2) stably transfected with mutated Harvey-RAS oncogeneCitation39 and other adenocarcinoma cell lines ( and ).
Although the above findings provide a genetic explanation for miR-21 overexpression based on RAS pathway deregulation, a non-negligible percentage of tumors expressing high miR-21 levels may not bear EGFR/RAS/BRAF mutations.Citation22,Citation30 To dissect this issue, we examined the contribution of epigenetic mechanisms to miR-21 deregulation. Indeed, in recent years it has become evident that hypermethylation of DNA CpG islands and aberrant histone posttranslational modifications on miRNA promoters might affect miRNAs expression.Citation50-Citation52 Furthermore, concerning the role of epigenetic mechanisms on miR-21 biogenesis, Ribas et al. have recently demonstrated a novel independent-regulated source of miR-21.Citation53 The miR-21 gene lies on vacuole membrane protein-1 (VMP1) genetic locus and the authors identified a VMP1-miR-21 fusion transcript that is overexpressed in several cell lines and differentially responds to specific stimuli, such as epigenetic modifying agents. While numerous studies have already demonstrated the involvement of DNA methylation in miRNA regulation,Citation54,Citation55 the contribution of histone marks is still poorly studied and the few available studies are mainly focused on histone modifications able to silence miRNA expression. For instance, Li et al. have demonstrated that silencing of miR-22 expression in acute lymphoblastic leukemia involves histone modification, such as accumulation of H3K27me3, independently of DNA methylation.Citation50 In our study, ChIP analysis revealed that marks of active transcription H3K3me3, H3K9–14ac, and H3K27ac were enhanced on the MIR-21 promoter in CRC cell lines with EMT phenotype; on the other hand, histone marks associated with gene repression, such as H3K27me3 and H3K9me2, were completely absent from the MIR-21 gene promoter. A similar analysis performed in Caco-2 cells, which lack invasive capacity, has shown reduced levels of H3K3me3, H3K9–14ac, and H3K27ac on the MIR-21 promoter. Using siRNA to transiently silence GCN5, one of the major histone acetyltransferase enzymes, we were able to reduce the expression of miR-21 precursor mRNA. Based on this evidence, we suggest that the binding affinity of different transcription factors on the MIR-21 promoter appears to be in part controlled by epigenetic mechanisms, such as the combination of active and repressive histone posttranslational modifications operated by histone modifier enzymes. Interestingly, we have recently proven that deregulation of histone modifier enzymes can be a consequence of oncogene transformation in colon carcinoma cell lines.Citation56
ITGβ4 is a novel miR-21 target regulating cell migration and EMT
miR-21 targets a number of cancer-related genes. Among them are PDCD4, PTEN, SPRY2, RECK, TIAM1, TIPM3, and a network of key tumor-suppressive pathways, suggesting that miR-21 is involved in many malignant properties, including migration and invasion.Citation43,Citation57-Citation61 We sought to identify new miR-21 targets that might explain the role of miR-21 in EMT. Our research revealed ITGβ4 as a new miR-21 target. Indeed, miR-21 is able to modulate ITGβ4 protein expression not only via repression of mRNA translation acting on UTR regions, but also through mRNA degradation. miR-21 binding site was identified in the coding sequence of ITGβ4 mRNA and inhibition of miR-21 resulted in increased ΙΤGβ4 mRNA expression. We also report, for the first time, a potential active role for the negative strand of miR-21 (miR-21*), which is thought to be degraded after miR-21 maturation, since three putative miR-21* binding sites were predicted on the coding sequence of ΙΤGβ4.
ITGα6β4, a heterodimer of ITGα6 with ITGβ4, is expressed only in polarized cells, such as colon epithelial cells.Citation35 Along with plectin, ITGα6β4 is the major structural protein of the hemidesmosome, a formation that anchors cells to the extracellular matrix.Citation62 ITGα6β4 was shown to be able to inhibit colon cancer cell proliferation and MYC activity.Citation63 Yu et al. showed that plectin, when associated with ITGβ4, serves to restrain pancreatic cancer cell migration, an interaction that is disrupted by RON signaling.Citation64 Similar to these findings, we observed a reduction of cell migration in anti-miR-21 treated cells, as a consequence, at least partly, of ITGβ4 upregulation. Notably, in all cell lines analyzed, the levels of ITGβ4 mRNA were not correlated with the respective protein levels implying a posttranscriptional regulation of this gene. We speculate that re-expression of ITGβ4, along with other factors, forms both strong cell-cell and cell-surface interactions that reduce the ability of cancer cells to move, as shown by confocal pictures. This was proved in the present study by coupling cell migration assay with gain of function experiments (overexpression of miR-21) and by a functional blocking of ITGβ4 protein in living cells.
ITGβ4/MIR-21/PDCD4 three-gene assay can predict metastatic potential of CRC
The potential implication of the three-gene, ITGβ4/MIR-21/PDCD4, deregulated expression in cancer cell movement was also studied in clinical CRC, by the analysis of association to local (regional lymph node) and/or distant metastasis. Although ITGβ4 expression did not show a unique trend as was the case for miR-21 and PDCD4, its downregulation in tumor tissue samples helped to isolate a group of patients in which relevant expression of the three genes miR-21, ITGβ4, and PDCD4 can significantly discriminate between metastatic and non-metastatic patients. Strong cell-cell interactions need to be disrupted prior to cancer cells initiating the complex metastatic cascade, although this condition is not enough to confer a metastatic phenotype and a plethora of factors are involved in cell-cell interactions. Considering the above, we think that downregulation of ITGβ4 in certain cancer patients is important during the loosening of cell junctions and that, together with other pro-metastatic factors (e.g., miR-21 and PTEN), regulates metastasis. On the other hand, for cancer patients where ITGβ4 expression did not change or was even increased, it can be hypothesized that the loosening of cell interactions, a step toward metastasis, depends on other unidentified factors. Nevertheless, liver metastasis is detected in up to 25% at primary diagnosis and occurs during disease course in 50–70% after 3 y of follow up. Surgical resection of liver is the only therapy that produces long-term cure in CRC patients with hepatic metastasis.Citation65,Citation66 Prognostic tools, based on molecular markers, that allow prediction of metastatic potential in the primary tumor tissue could improve quality of pathological reports and allow appropriate selection of patients that need special care and follow up. Our findings also enlarge the cell-biology pathways that, under the control of microRNAs, regulate the metastatic process. In fact, it is established that microRNAs, through specific regulatory networks, control metastatic progression both via cell-intrinsic and cell-extrinsic networks.Citation67
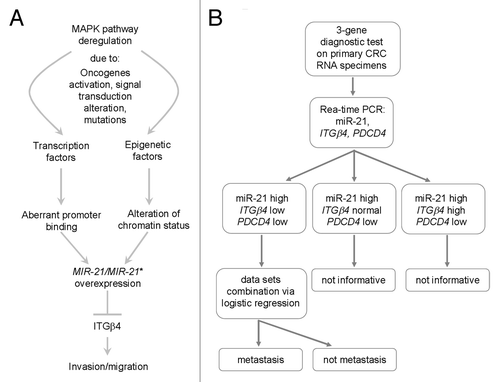
Considering all results, we suggest an interesting network that probably controls cell migration and EMT in CRC cell lines and a potential diagnostic tool based on miR-21- and targets-expression (). Epigenetic and genetic pathways, subsequent to initial carcinogenic stimuli, induce in parallel transcriptional deregulation of critical cancer-associated genes and loci. Among them, miR-21 gene causes downregulation of tumor suppressor genes (e.g., PDCD4) and epithelial key genes, such as ITGβ4, enhancing malignant transformation and EMT. Clinical data could be further exploited, since relevant expression of miR-21, ITGβ4, and PDCD4 in a subgroup of specimens can significantly discriminate between patients with and without metastasis. Finally, considering the ITGβ4 inactivating experiments (), putative CRC anti-metastatic therapies, aimed to inhibit integrins function, should be carefully administrated since they might not suppress cell migration but paradoxically activate cancer cell locomotion.
Figure 8. Proposed model of miR-21 regulation and action in cancer and 3-gene diagnostic test. (A) A variety of stimuli in tumor tissue might activate genetic (e.g., transcription factors) and epigenetic (e.g., histone modifier enzymes) factors leading to aberrant gene expression of miR-21. As consequence, key epithelial genes such as ITGβ4 are repressed and the invasive and metastatic capacity of cancer cells is augmented, also by regulating EMT. (B) Flowchart of 3-gene diagnostic test performed on CRC RNA specimens coming from primary tumors.
Materials and Methods
Cell culture
Caco-2, DLD-1, HT-29, SW620, HCT116, Colo 205, RKO cell lines were maintained in DMEM medium, containing 10% fetal bovine serum (FBS), 1X penicillin, 1X streptomycin, and 2 mM L-glutamine. Cells were incubated at 37 °C, 5% CO2 in humidified atmosphere.
Migration and invasion assays
Cells were trypsinized, washed with medium containing 1% FBS, and counted. 105 cells were plated into upper chamber of an 8μm-pore Transwell filter (Corning), mounted in a 24-well dish containing 10% FBS medium. Filters were pre-coated with fibronectin. Cells were allowed to migrate at 37 °C, 5% CO2 for 36–40 h, fixed with methanol and stained with 0.1% w/v crystal violet. For migration assay in presence of ITGβ4 blocking antibody, a final concentration of 50 ng/ml of IgG was used both for anti-ITGβ4 and for control antibody. Underside of filters were observed with 40× objective and migrating cells were determined in each well. Experiments were performed in duplicate and repeated twice.
Plasmid DNA (pDNA), small interfering RNA (siRNA), miRNA mimic and miRNA inhibitor transfections
For pDNA transfection, Caco-2 cells were transfected with 2 μg of plasmid DNA in a 6-well plate using Lipofectamine™ 2000 (Invitrogen, USA). For siRNA transfection, cells were seeded in 6-well plates and transfected with 50–100 nM of ON-TARGETplus SMARTpool (Dharmacon) according to the manufacturer’s instructions.
For miRNA mimic and inhibitor transfection, cells were seeded in 6-well plates and transfected with 10–20 pmol of miRCURY LNA™ microRNA Inhibitor (Exiqon) or with 90 pmol of mirVana miRNA mimic (Ambion) according to the manufacturer’s instructions. Protein and RNA extracts have been prepared 24/48 h after transfection.
RNA extraction and reverse transcription
Total RNA was extracted using the TRI®Reagent (Invitrogen) following the manufacturer’s instructions. First-strand cDNA was synthesized using the Superscript™ II Reverse Transcriptase (Invitrogen), according to the manufacturer’s instructions. The reaction mixture contained 2 μg total RNA diluted water. The final reaction volume was 20 μL.
Quantitative PCR
Quantitative PCR was performed using the SYBR® Green chemistry, in 96-well plates (Bio-Rad). PCR runs and fluorescence detection were performed in an iQ™5 Detection System (Bio-Rad). The reaction mixture (10 μl) contained 50 ng of cDNA, 5 μL Supermix (Bio-Rad), and 140 nM of gene-specific primers. The reaction conditions were: 95 °C for 10 min, 40 cycles of 95 °C for 15 sec and 60 °C for 60 sec. Each reaction was performed in triplicate. Dissociation curves of PCR products were generated after amplification. Primer sequences are listed in Table S2. For mature MIR21 quantification in relation to RNU6B expression, reverse transcription of 10 ng total RNA was performed using the MicroRNA Reverse Transcription Kit, following the manufacturer’s instructions. Quantitative PCR was performed using the TaqMan® chemistry (Applied-Biosystems). Experiments have been performed in duplicate and repeated twice.
Chromatin immunoprecipitation (ChIP)
Pre-cleared chromatin was incubated overnight by rotation with 3–4 μg of antibody or no antibody as a negative control. Two μL of the immunoprecipitated DNA were used to evaluate miR-21 promoter enrichment by real-time PCR. The results were normalized against levels of an unrelated sequence lying within the coding sequence of the GAPDH. Primer sequences and antibody are listed in Tables S2 and S3 respectively. Experiments have been performed in duplicate and repeated twice.
Western blotting
Total protein was extracted with 60 μL RIPA lysis buffer and western blotting was performed according to standard protocols. Blots were incubated overnight at 4 °C with the appropriate primary antibodies. Antibodies used are listed in Table S2. Protein extracts have been prepared for at least two independent experiments and blots repeated at least twice.
Immunofluorescence assays
Cells used for immunofluorescence assays were fixed with methanol/acetone solution (8:1), washed, permeabilized with 0.3% Triton-X-100, and blocked with 5% BSA prior to incubation with primary antibodies. Nuclei were counterstained with Hoechst.
Clinical specimens
All patients were informed and gave written informed consent. The study protocol was approved by the Medical Ethical Review Committee of G. Genimatas General Hospital of Athens.
| Abbreviations: | ||
| CRC | = | colorectal cancer |
| EMT | = | epithelial-mesenchymal transition |
| H3K9me3 | = | trimethyl histone H3 lysine 9 |
| H3K27me3 | = | trimethyl histone H3 lysine 27 |
| PI3K | = | phosphatidylinositol 3-kinase |
| ITGB4 | = | Integrin β4 |
| ChIP | = | chromatin immunoprecipitation |
| PDCD4 | = | Programmed cell death 4 |
Additional material
Download Zip (836.3 KB)Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
This study was funded by EU FP7 Project 241783 EpiDiaCan and support by GSRT to Pintzas A.
Supplemental Materials
Supplemental materials may be found here: www.landesbioscience.com/journals/epigenetics/article/26842
References
- Steeg PS. Tumor metastasis: mechanistic insights and clinical challenges. Nat Med 2006; 12:895 - 904; http://dx.doi.org/10.1038/nm1469; PMID: 16892035
- Thiery JP. Epithelial-mesenchymal transitions in tumour progression. Nat Rev Cancer 2002; 2:442 - 54; http://dx.doi.org/10.1038/nrc822; PMID: 12189386
- Voulgari A, Pintzas A. Epithelial-mesenchymal transition in cancer metastasis: mechanisms, markers and strategies to overcome drug resistance in the clinic. Biochim Biophys Acta 2009; 1796:75 - 90; PMID: 19306912
- Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer 2006; 6:857 - 66; http://dx.doi.org/10.1038/nrc1997; PMID: 17060945
- Deng S, Calin GA, Croce CM, Coukos G, Zhang L. Mechanisms of microRNA deregulation in human cancer. Cell Cycle 2008; 7:2643 - 6; http://dx.doi.org/10.4161/cc.7.17.6597; PMID: 18719391
- Mani SA, Yang J, Brooks M, Schwaninger G, Zhou A, Miura N, Kutok JL, Hartwell K, Richardson AL, Weinberg RA. Mesenchyme Forkhead 1 (FOXC2) plays a key role in metastasis and is associated with aggressive basal-like breast cancers. Proc Natl Acad Sci U S A 2007; 104:10069 - 74; http://dx.doi.org/10.1073/pnas.0703900104; PMID: 17537911
- Yang J, Mani SA, Donaher JL, Ramaswamy S, Itzykson RA, Come C, Savagner P, Gitelman I, Richardson A, Weinberg RA. Twist, a master regulator of morphogenesis, plays an essential role in tumor metastasis. Cell 2004; 117:927 - 39; http://dx.doi.org/10.1016/j.cell.2004.06.006; PMID: 15210113
- Cano A, Pérez-Moreno MA, Rodrigo I, Locascio A, Blanco MJ, del Barrio MG, Portillo F, Nieto MA. The transcription factor snail controls epithelial-mesenchymal transitions by repressing E-cadherin expression. Nat Cell Biol 2000; 2:76 - 83; http://dx.doi.org/10.1038/35000025; PMID: 10655586
- Shaulian E, Karin M. AP-1 as a regulator of cell life and death. Nat Cell Biol 2002; 4:E131 - 6; http://dx.doi.org/10.1038/ncb0502-e131; PMID: 11988758
- Ozanne BW, Spence HJ, McGarry LC, Hennigan RF. Transcription factors control invasion: AP-1 the first among equals. Oncogene 2007; 26:1 - 10; http://dx.doi.org/10.1038/sj.onc.1209759; PMID: 16799638
- Eferl R, Wagner EF. AP-1: a double-edged sword in tumorigenesis. Nat Rev Cancer 2003; 3:859 - 68; http://dx.doi.org/10.1038/nrc1209; PMID: 14668816
- Mechta F, Lallemand D, Pfarr CM, Yaniv M. Transformation by ras modifies AP1 composition and activity. Oncogene 1997; 14:837 - 47; http://dx.doi.org/10.1038/sj.onc.1200900; PMID: 9047391
- Rajalingam K, Schreck R, Rapp UR, Albert S. Ras oncogenes and their downstream targets. Biochim Biophys Acta 2007; 1773:1177 - 95; http://dx.doi.org/10.1016/j.bbamcr.2007.01.012; PMID: 17428555
- Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca FA, Visone R, Iorio M, Roldo C, Ferracin M, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci U S A 2006; 103:2257 - 61; http://dx.doi.org/10.1073/pnas.0510565103; PMID: 16461460
- Iorio MV, Ferracin M, Liu CG, Veronese A, Spizzo R, Sabbioni S, Magri E, Pedriali M, Fabbri M, Campiglio M, et al. MicroRNA gene expression deregulation in human breast cancer. Cancer Res 2005; 65:7065 - 70; http://dx.doi.org/10.1158/0008-5472.CAN-05-1783; PMID: 16103053
- Ribas J, Ni X, Haffner M, Wentzel EA, Salmasi AH, Chowdhury WH, Kudrolli TA, Yegnasubramanian S, Luo J, Rodriguez R, et al. miR-21: an androgen receptor-regulated microRNA that promotes hormone-dependent and hormone-independent prostate cancer growth. Cancer Res 2009; 69:7165 - 9; http://dx.doi.org/10.1158/0008-5472.CAN-09-1448; PMID: 19738047
- Schetter AJ, Leung SY, Sohn JJ, Zanetti KA, Bowman ED, Yanaihara N, Yuen ST, Chan TL, Kwong DL, Au GK, et al. MicroRNA expression profiles associated with prognosis and therapeutic outcome in colon adenocarcinoma. JAMA 2008; 299:425 - 36; http://dx.doi.org/10.1001/jama.299.4.425; PMID: 18230780
- Markou A, Tsaroucha EG, Kaklamanis L, Fotinou M, Georgoulias V, Lianidou ES. Prognostic value of mature microRNA-21 and microRNA-205 overexpression in non-small cell lung cancer by quantitative real-time RT-PCR. Clin Chem 2008; 54:1696 - 704; http://dx.doi.org/10.1373/clinchem.2007.101741; PMID: 18719201
- Chan JA, Krichevsky AM, Kosik KS. MicroRNA-21 is an antiapoptotic factor in human glioblastoma cells. Cancer Res 2005; 65:6029 - 33; http://dx.doi.org/10.1158/0008-5472.CAN-05-0137; PMID: 16024602
- Kjaer-Frifeldt S, Hansen TF, Nielsen BS, Joergensen S, Lindebjerg J, Soerensen FB, dePont Christensen R, Jakobsen A, Danish Colorectal Cancer Group. The prognostic importance of miR-21 in stage II colon cancer: a population-based study. Br J Cancer 2012; 107:1169 - 74; http://dx.doi.org/10.1038/bjc.2012.365; PMID: 23011541
- Nielsen BS, Jørgensen S, Fog JU, Søkilde R, Christensen IJ, Hansen U, Brünner N, Baker A, Møller S, Nielsen HJ. High levels of microRNA-21 in the stroma of colorectal cancers predict short disease-free survival in stage II colon cancer patients. Clin Exp Metastasis 2011; 28:27 - 38; http://dx.doi.org/10.1007/s10585-010-9355-7; PMID: 21069438
- Talotta F, Cimmino A, Matarazzo MR, Casalino L, De Vita G, D’Esposito M, Di Lauro R, Verde P. An autoregulatory loop mediated by miR-21 and PDCD4 controls the AP-1 activity in RAS transformation. Oncogene 2009; 28:73 - 84; http://dx.doi.org/10.1038/onc.2008.370; PMID: 18850008
- Hatley ME, Patrick DM, Garcia MR, Richardson JA, Bassel-Duby R, van Rooij E, Olson EN. Modulation of K-Ras-dependent lung tumorigenesis by MicroRNA-21. Cancer Cell 2010; 18:282 - 93; http://dx.doi.org/10.1016/j.ccr.2010.08.013; PMID: 20832755
- Yang N, Coukos G, Zhang L. MicroRNA epigenetic alterations in human cancer: one step forward in diagnosis and treatment. Int J Cancer 2008; 122:963 - 8; http://dx.doi.org/10.1002/ijc.23325; PMID: 18098137
- Saito Y, Liang G, Egger G, Friedman JM, Chuang JC, Coetzee GA, Jones PA. Specific activation of microRNA-127 with downregulation of the proto-oncogene BCL6 by chromatin-modifying drugs in human cancer cells. Cancer Cell 2006; 9:435 - 43; http://dx.doi.org/10.1016/j.ccr.2006.04.020; PMID: 16766263
- Calin GA, Ferracin M, Cimmino A, Di Leva G, Shimizu M, Wojcik SE, Iorio MV, Visone R, Sever NI, Fabbri M, et al. A MicroRNA signature associated with prognosis and progression in chronic lymphocytic leukemia. N Engl J Med 2005; 353:1793 - 801; http://dx.doi.org/10.1056/NEJMoa050995; PMID: 16251535
- Zhang L, Volinia S, Bonome T, Calin GA, Greshock J, Yang N, Liu CG, Giannakakis A, Alexiou P, Hasegawa K, et al. Genomic and epigenetic alterations deregulate microRNA expression in human epithelial ovarian cancer. Proc Natl Acad Sci U S A 2008; 105:7004 - 9; http://dx.doi.org/10.1073/pnas.0801615105; PMID: 18458333
- Thomson JM, Newman M, Parker JS, Morin-Kensicki EM, Wright T, Hammond SM. Extensive post-transcriptional regulation of microRNAs and its implications for cancer. Genes Dev 2006; 20:2202 - 7; http://dx.doi.org/10.1101/gad.1444406; PMID: 16882971
- Iliopoulos D, Jaeger SA, Hirsch HA, Bulyk ML, Struhl K. STAT3 activation of miR-21 and miR-181b-1 via PTEN and CYLD are part of the epigenetic switch linking inflammation to cancer. Mol Cell 2010; 39:493 - 506; http://dx.doi.org/10.1016/j.molcel.2010.07.023; PMID: 20797623
- Iliopoulos D, Hirsch HA, Struhl K. An epigenetic switch involving NF-kappaB, Lin28, Let-7 MicroRNA, and IL6 links inflammation to cell transformation. Cell 2009; 139:693 - 706; http://dx.doi.org/10.1016/j.cell.2009.10.014; PMID: 19878981
- Kouzarides T. Chromatin modifications and their function. Cell 2007; 128:693 - 705; http://dx.doi.org/10.1016/j.cell.2007.02.005; PMID: 17320507
- Chi P, Allis CD, Wang GG. Covalent histone modifications--miswritten, misinterpreted and mis-erased in human cancers. Nat Rev Cancer 2010; 10:457 - 69; http://dx.doi.org/10.1038/nrc2876; PMID: 20574448
- Beaulieu JF. Extracellular matrix components and integrins in relationship to human intestinal epithelial cell differentiation. Prog Histochem Cytochem 1997; 31:1 - 78; http://dx.doi.org/10.1016/S0079-6336(97)80001-0; PMID: 9088045
- Teller IC, Beaulieu JF. Interactions between laminin and epithelial cells in intestinal health and disease. Expert Rev Mol Med 2001; 3:1 - 18; http://dx.doi.org/10.1017/S1462399401003623; PMID: 14585148
- Beaulieu JF. Integrin α6β4 in colorectal cancer. World J Gastrointest Pathophysiol 2010; 1:3 - 11; PMID: 21607137
- Stallmach A, von Lampe B, Matthes H, Bornhöft G, Riecken EO. Diminished expression of integrin adhesion molecules on human colonic epithelial cells during the benign to malign tumour transformation. Gut 1992; 33:342 - 6; http://dx.doi.org/10.1136/gut.33.3.342; PMID: 1568652
- Falcioni R, Turchi V, Vitullo P, Navarra G, Ficari F, Cavaliere F, Sacchi A, Marianicostantini R. Integrin Beta-4 expression in colorectal-cancer. Int J Oncol 1994; 5:573 - 8; PMID: 21559615
- Sordat I, Bosman FT, Dorta G, Rousselle P, Aberdam D, Blum AL, Sordat B. Differential expression of laminin-5 subunits and integrin receptors in human colorectal neoplasia. J Pathol 1998; 185:44 - 52; http://dx.doi.org/10.1002/(SICI)1096-9896(199805)185:1<44::AID-PATH46>3.0.CO;2-A; PMID: 9713359
- Roberts ML, Drosopoulos KG, Vasileiou I, Stricker M, Taoufik E, Maercker C, Guialis A, Alexis MN, Pintzas A. Microarray analysis of the differential transformation mediated by Kirsten and Harvey Ras oncogenes in a human colorectal adenocarcinoma cell line. Int J Cancer 2006; 118:616 - 27; http://dx.doi.org/10.1002/ijc.21386; PMID: 16152623
- Asangani IA, Rasheed SA, Nikolova DA, Leupold JH, Colburn NH, Post S, Allgayer H. MicroRNA-21 (miR-21) post-transcriptionally downregulates tumor suppressor Pdcd4 and stimulates invasion, intravasation and metastasis in colorectal cancer. Oncogene 2008; 27:2128 - 36; http://dx.doi.org/10.1038/sj.onc.1210856; PMID: 17968323
- Fujita S, Ito T, Mizutani T, Minoguchi S, Yamamichi N, Sakurai K, Iba H. miR-21 Gene expression triggered by AP-1 is sustained through a double-negative feedback mechanism. J Mol Biol 2008; 378:492 - 504; http://dx.doi.org/10.1016/j.jmb.2008.03.015; PMID: 18384814
- Huang HY, Chien CH, Jen KH, Huang HD. RegRNA: an integrated web server for identifying regulatory RNA motifs and elements. Nucleic Acids Res 2006; 34:Web Server issue W429-34; http://dx.doi.org/10.1093/nar/gkl333; PMID: 16845041
- Fassan M, Pizzi M, Giacomelli L, Mescoli C, Ludwig K, Pucciarelli S, Rugge M. PDCD4 nuclear loss inversely correlates with miR-21 levels in colon carcinogenesis. Virchows Arch 2011; 458:413 - 9; http://dx.doi.org/10.1007/s00428-011-1046-5; PMID: 21279518
- Shibuya H, Iinuma H, Shimada R, Horiuchi A, Watanabe T. Clinicopathological and prognostic value of microRNA-21 and microRNA-155 in colorectal cancer. Oncology 2010; 79:313 - 20; http://dx.doi.org/10.1159/000323283; PMID: 21412018
- Vaiopoulos AG, Papachroni KK, Papavassiliou AG. Colon carcinogenesis: Learning from NF-kappaB and AP-1. Int J Biochem Cell Biol 2010; 42:1061 - 5; http://dx.doi.org/10.1016/j.biocel.2010.03.018; PMID: 20348011
- Sayed D, He M, Hong C, Gao S, Rane S, Yang Z, Abdellatif M. MicroRNA-21 is a downstream effector of AKT that mediates its antiapoptotic effects via suppression of Fas ligand. J Biol Chem 2010; 285:20281 - 90; http://dx.doi.org/10.1074/jbc.M110.109207; PMID: 20404348
- Bandres E, Agirre X, Bitarte N, Ramirez N, Zarate R, Roman-Gomez J, Prosper F, Garcia-Foncillas J. Epigenetic regulation of microRNA expression in colorectal cancer. Int J Cancer 2009; 125:2737 - 43; http://dx.doi.org/10.1002/ijc.24638; PMID: 19521961
- Belguise K, Kersual N, Galtier F, Chalbos D. FRA-1 expression level regulates proliferation and invasiveness of breast cancer cells. Oncogene 2005; 24:1434 - 44; http://dx.doi.org/10.1038/sj.onc.1208312; PMID: 15608675
- Chiappetta G, Ferraro A, Botti G, Monaco M, Pasquinelli R, Vuttariello E, Arnaldi L, Di Bonito M, D’Aiuto G, Pierantoni GM, et al. FRA-1 protein overexpression is a feature of hyperplastic and neoplastic breast disorders. BMC Cancer 2007; 7:17; http://dx.doi.org/10.1186/1471-2407-7-17; PMID: 17254320
- Li X, Liu J, Zhou R, Huang S, Huang S, Chen XM. Gene silencing of MIR22 in acute lymphoblastic leukaemia involves histone modifications independent of promoter DNA methylation. Br J Haematol 2010; 148:69 - 79; http://dx.doi.org/10.1111/j.1365-2141.2009.07920.x; PMID: 19807731
- Zhang Z, Zhang MQ. Histone modification profiles are predictive for tissue/cell-type specific expression of both protein-coding and microRNA genes. BMC Bioinformatics 2011; 12:155; http://dx.doi.org/10.1186/1471-2105-12-155; PMID: 21569556
- Suzuki H, Takatsuka S, Akashi H, Yamamoto E, Nojima M, Maruyama R, Kai M, Yamano HO, Sasaki Y, Tokino T, et al. Genome-wide profiling of chromatin signatures reveals epigenetic regulation of MicroRNA genes in colorectal cancer. Cancer Res 2011; 71:5646 - 58; http://dx.doi.org/10.1158/0008-5472.CAN-11-1076; PMID: 21734013
- Ribas J, Ni X, Castanares M, Liu MM, Esopi D, Yegnasubramanian S, Rodriguez R, Mendell JT, Lupold SE. A novel source for miR-21 expression through the alternative polyadenylation of VMP1 gene transcripts. Nucleic Acids Res 2012; 40:6821 - 33; http://dx.doi.org/10.1093/nar/gks308; PMID: 22505577
- Lin PC, Chiu YL, Banerjee S, Park K, Mosquera JM, Giannopoulou E, Alves P, Tewari AK, Gerstein MB, Beltran H, et al. Epigenetic repression of miR-31 disrupts androgen receptor homeostasis and contributes to prostate cancer progression. Cancer Res 2013; 73:1232 - 44; http://dx.doi.org/10.1158/0008-5472.CAN-12-2968; PMID: 23233736
- Baer C, Claus R, Plass C. Genome-wide epigenetic regulation of miRNAs in cancer. Cancer Res 2013; 73:473 - 7; http://dx.doi.org/10.1158/0008-5472.CAN-12-3731; PMID: 23316035
- Peláez IM, Kalogeropoulou M, Ferraro A, Voulgari A, Pankotai T, Boros I, Pintzas A. Oncogenic RAS alters the global and gene-specific histone modification pattern during epithelial-mesenchymal transition in colorectal carcinoma cells. Int J Biochem Cell Biol 2010; 42:911 - 20; http://dx.doi.org/10.1016/j.biocel.2010.01.024; PMID: 20109579
- Meng F, Henson R, Wehbe-Janek H, Ghoshal K, Jacob ST, Patel T. MicroRNA-21 regulates expression of the PTEN tumor suppressor gene in human hepatocellular cancer. Gastroenterology 2007; 133:647 - 58; http://dx.doi.org/10.1053/j.gastro.2007.05.022; PMID: 17681183
- Mei Y, Bian C, Li J, Du Z, Zhou H, Yang Z, Zhao RC. miR-21 modulates the ERK-MAPK signaling pathway by regulating SPRY2 expression during human mesenchymal stem cell differentiation. J Cell Biochem 2013; 114:1374 - 84; http://dx.doi.org/10.1002/jcb.24479; PMID: 23239100
- Cottonham CL, Kaneko S, Xu L. miR-21 and miR-31 converge on TIAM1 to regulate migration and invasion of colon carcinoma cells. J Biol Chem 2010; 285:35293 - 302; http://dx.doi.org/10.1074/jbc.M110.160069; PMID: 20826792
- Gabriely G, Wurdinger T, Kesari S, Esau CC, Burchard J, Linsley PS, Krichevsky AM. MicroRNA 21 promotes glioma invasion by targeting matrix metalloproteinase regulators. Mol Cell Biol 2008; 28:5369 - 80; http://dx.doi.org/10.1128/MCB.00479-08; PMID: 18591254
- Papagiannakopoulos T, Shapiro A, Kosik KS. MicroRNA-21 targets a network of key tumor-suppressive pathways in glioblastoma cells. Cancer Res 2008; 68:8164 - 72; http://dx.doi.org/10.1158/0008-5472.CAN-08-1305; PMID: 18829576
- Leung CL, Green KJ, Liem RK. Plakins: a family of versatile cytolinker proteins. Trends Cell Biol 2002; 12:37 - 45; http://dx.doi.org/10.1016/S0962-8924(01)02180-8; PMID: 11854008
- Dydensborg AB, Teller IC, Groulx JF, Basora N, Paré F, Herring E, Gauthier R, Jean D, Beaulieu JF. Integrin alpha6Bbeta4 inhibits colon cancer cell proliferation and c-Myc activity. BMC Cancer 2009; 9:223; http://dx.doi.org/10.1186/1471-2407-9-223; PMID: 19586553
- Yu PT, Babicky M, Jaquish D, French R, Marayuma K, Mose E, Niessen S, Hoover H, Shields D, Cheresh D, et al. The RON-receptor regulates pancreatic cancer cell migration through phosphorylation-dependent breakdown of the hemidesmosome. Int J Cancer 2012; 131:1744 - 54; http://dx.doi.org/10.1002/ijc.27447; PMID: 22275185
- Ries LAG, Melbert D, Krapcho M, Mariotto A, Miller BA, Feuer EJ, Clegg L, Horner MJ, Howlader N, Eisner MP, et al., eds. SEER Cancer Statistics Review, 1975-2004, National Cancer Institute. Bethesda, MD. Available at http://seer.cancer.gov/csr/1975_2004/
- Nordlinger B, Rougier P. Liver metastases from colorectal cancer: the turning point. J Clin Oncol 2002; 20:1442 - 5; PMID: 11896089
- Pencheva N, Tavazoie SF. Control of metastatic progression by microRNA regulatory networks. Nat Cell Biol 2013; 15:546 - 54; http://dx.doi.org/10.1038/ncb2769; PMID: 23728460
