Abstract
DNA methylation and polycomb proteins are well-known mediators of epigenetic silencing in mammalian cells. Usually described as mutually exclusive, this statement is today controversial and recent in vitro studies suggest the co-existence of both repressor systems. We addressed this issue in the study of Retinoic Acid Receptor β (RARβ), a tumor suppressor gene frequently silenced in prostate cancer. We found that the RARβ promoter is hypermethylated in all studied prostate tumors and methylation levels are positively correlated with H3K27me3 enrichments. Thus, by using bisulfite conversion and pyrosequencing of immunoprecipitated H3K27me3 chromatin, we demonstrated that DNA methylation and polycomb repression co-exist in vivo at this locus. We found this repressive association in 6/6 patient tumor samples of different Gleason score, suggesting a strong interplay of DNA methylation and EZH2 to silence RARβ during prostate tumorigenesis.
Introduction
Epigenetic repression is a common cellular mechanism to silence gene expression. To this goal, two major actors are DNA methylation and polycomb proteins. While these two repressing systems are well described, the interplay between them is still a key question in cellular biology in order to better understand both cell differentiation and tumorigenesis. Several reports have shown that DNA methylation and H3K27me3, catalyzed by EZH2 as part of the Polycomb Repressive Complex 2 (PRC2), are mutually exclusive.Citation1-Citation3 Brinkman et al. addressed the overlap of these two marks at the genome-wide scale using ChIP-bisulfite-sequencing (ChIP-BS-seq) on the HCT116 tumor cell line and mouse embryonic stem (ES) cells.Citation4 This work concluded that DNA methylation and H3K27me3 can co-exist, except for CpG-dense regions where they are mutually exclusive. Meanwhile, with the same experimental strategy, Statham et al. described that in normal and prostate cancer cells DNA methylation and H3K27me3 are not always mutually exclusive and that they can co-occur according to the genomic context. Further, they showed for the first time that silenced genes with CpG island promoters can display both epigenetic silencing marks.Citation5
In parallel to these studies we have characterized the two marks, DNA methylation and H3K27me3, on the Retinoic Acid Receptor Beta (RARβ) promoter in prostate cancer cell lines.Citation6 RARβ gene encodes several isoforms from two distinct promoters named P1 and P2. Among them, the RARβ2 isoform is transcribed from the CpG-rich RARβ P2 promoter and act as a tumor suppressor. Its expression is frequently silenced leading to treatment resistance by Retinoic Acid (RA).Citation7,Citation8 In our recent study, we observed and characterized distinct epigenetic silencing profiles at the RARβ P2 promoter in three prostate cancer cell lines: DU145 cells showed moderate DNA methylation concomitant with the H3K27me3 mark, while LNCaP and VCaP cells displayed high DNA methylation levels and no H3K27me3.Citation6
It has been shown that, in specific cases, DNA methylation and polycomb proteins may cooperate to repress genes. For example, target genes of polycomb proteins are known to be 12 times more likely to undergo DNA hypermethylation during tumorigenesisCitation9-Citation11 and polycomb proteins may directly interact with DNA methyltransferases (DNMTs).Citation12,Citation13 Because of their involvement in cancer, it is of great interest to understand how their deregulation leads to inappropriate gene silencing and the above cited contradictory reports require more investigation to clarify the crosstalk between DNA methylation and polycomb repression, particularly in biologically relevant material such as tumor specimens. Here we studied this interplay at the RARβ locus in six prostate tumor samples. We focused on the P2 promoter containing a CpG island and driving RARβ2 tumor suppressor expression (referred as RARβ2 promoter herein).
Results and Discussion
RARβ2 is a tumor suppressor gene frequently hypermethylated during breast and prostate tumorigenesis.Citation14-Citation17 Previously, we reported its hypermethylation in four prostate tumor cell lines (DU145, PC3, LNCaP and VCaP) and found that RARβ2 promoter hypermethylation is associated with H3K27me3 in DU145 prostate cancer cells but not in LNCaP and VCaP cells.Citation6
The relevance of studies using in vitro cell lines is always of debate, especially in the epigenetic field, as microenvironment and culture conditions differ dramatically from in vivo conditions and can modify the chromatin profile of cells.Citation18 Thus, we investigated the chromatin patterns of the RARβ2 promoter in six human prostate tumors (T1 to T6) with various Gleason scores. We analyzed the RARβ2 DNA methylation level in each tumor by bisulfite-pyrosequencing of 10 CpGs located in the gene promoter (downstream of the TSS, between +1 and +100). All of the tumors displayed RARβ2 promoter hypermethylation with average levels ranging from 34.9% to 72.7% (; Table S1). In comparison, the non-malignant cell line named EPT2,Citation19,Citation20 derived from primary prostate epithelial cells, showed a 2.4% DNA methylation level. This finding in our cohort of six patient tumors, representing low- and high-grade cancers (Gleason score from 5 to 9, ), confirmed that RARβ2 CpG island is targeted by DNA methylation in prostate cancer. Noteworthy, RARβ2 methylation levels are not correlated with patient age (Pearson’s r = -0.35) nor with tumor grade (Pearson’s r = 0.47) in our samples.
Table 1.RARβ2 promoter is hypermethylated in six human prostate tumors
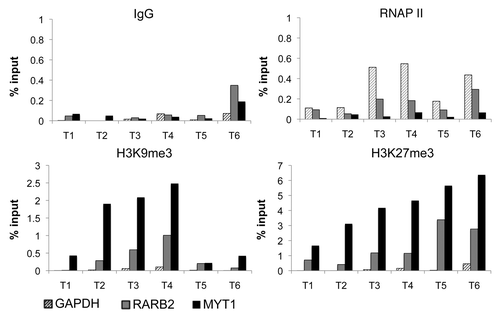
We then investigated the histone marks associated with the RARβ2 promoter hypermethylation in these six prostate cancer samples (). Using small amounts of fresh-frozen tumor samples, we performed ChIP experiments to detect the heterochromatin H3K9 trimethylation mark (H3K9me3) and the polycomb H3K27me3 mark. As negative control, we chose the GAPDH promoter at which these two repressive marks were absent, while RNA polymerase II (RNAP II) was enriched, consistently with the expression of this housekeeping gene (, hatched bars). MYT1 promoter has been previously reported as repressed by polycomb proteinsCitation21 and thus was here used as positive control for the polycomb mark H3K27me3. Accordingly, we detected a strong enrichment of H3K27me3 together with variable levels of H3K9me3 and absence of RNAP II (, black bars). Interestingly, at the RARβ2 promoter, the H3K27me3 repressive mark was present in all tumor samples and H3K9me3 was detected in four of them (, gray bars). The weak enrichment for RNAP II measured in some of the samples might suggests residual low expression of the gene. These results demonstrate that repressive histone marks are associated with hypermethylated RARβ2 promoter, and in particular with the polycomb mark H3K27me3. Indeed, we found a positive correlation between RARβ2 promoter methylation levels and H3K27me3 enrichments (Pearson’s r = 0.84), while no such correlation is observed with H3K9me3 (Pearson’s r = 0.15). These results suggest that an interplay between DNA methylation and polycomb repression occur at RARβ2 CpG-rich locus in prostate tumorigenesis. For comparison, in the EPT2 cell line derived from primary prostate epithelial cells, the non-methylated RARβ2 promoter did not display enrichment for H3K27me3 mark (data not shown).
Figure 1. Repressive chromatin pattern at the RARβ2 promoter in prostate cancer. ChIP analysis of RNAP II, H3K9me3 and H3K27me3 enrichment in six prostate tumors (T1-T6). IgG is used as control to measure non-specific immunoprecipitation. Enrichments were analyzed by qPCR at the RARβ2, GAPDH and MYT1 promoters.
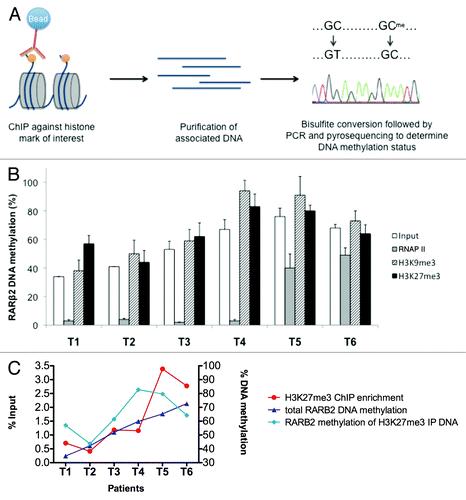
To directly assess the co-existence of DNA methylation and H3K27me3 histone mark on the same DNA molecule, bisulfite conversion and pyrosequencing was performed on the immunoprecipitated ChIP samples analyzed in . ChIP-bisulfite-pyrosequencing (ChIP-BS-pyro) allowed us to measure the DNA methylation level associated with a chromatin modification of interest (). As expected, ChIP input samples T1 to T6 showed a similar RARβ2 methylation level () to that performed directly on genomic DNA () (Pearson’s r = 0.94). H3K9me3 immunoprecipitated samples showed levels of methylation equal or higher to input controls, confirming that H3K9me3 mark is tightly associated with DNA methylation. Remarkably, similar results were also observed in the H3K27me3 immunoprecipitated samples—and not in the RNAP II one—demonstrating that DNA methylation and polycomb repression co-exist at the same locus in tumor patients’ samples (). H3K9me3 and DNA methylation co-exist at the RARβ2 locus but no correlation exist between these two marks in the six tumors (Pearson’s r = 0.15). On the contrary, H3K27me3 co-exist with DNA methylation at the promoter and higher H3K27me3 ChIP enrichments are associated to higher DNA methylation with a positive correlation (Pearson’s r = 0.84) ().
Figure 2. Co-existence of H3K27me3 and DNA methylation at the RARβ2 promoter. (A) Schematic representation of the ChIP-BS-pyro experimental procedure. (B) Average percentage of RARβ2 promoter methylation measured by bisulfite conversion-pyrosequencing on input samples, RNAP II, H3K9me3 and H3K27me3 immunoprecipitated DNA. Results are presented for samples named T1 to T6 as mean +/− SD of PCR technical replicates. (C) Graphic representation of H3K27me3 ChIP enrichments at the RARβ2 promoter with RARβ2 methylation levels on total DNA and after H3K27me3 immunoprecipitation (IP) in the six patient samples (expressed as average methylation percentage).
All together, these results in patients’ samples are in agreement with our previous observations in the DU145 cell lineCitation6 and support in vitro data showing that these two silencing marks are not mutually exclusive at CpG islands.Citation5 Moreover, our findings at the RARβ2 locus strongly suggest an interplay between DNA hypermethylation and the polycomb mark H3K27me3 during prostate tumorigenesis.
Understanding of the crosstalk between DNA methylation and polycomb repression is an important issue in both normal and cancer development. Further studies are required to address how methylation and polycomb profiles are established and cooperate to achieve abnormal silencing of specific tumor suppressor gene. In human embryonic fibroblasts, RARβ2 has been shown to be a polycomb target geneCitation22 and polycomb proteins are deregulated during prostate tumorigenesis,Citation23 which could explain aberrant H3K27me3 tagging. Non-coding RNAs (ncRNA) could also play a part in recruiting polycomb to specific locus. We did not, however, identify ncRNAs associated with the RARβ2 promoter in prostate cancer cell lines.Citation24 Interestingly, Zhao et al.Citation25 identified a PRC2-interacting antisense transcript to the 3′ of RARβ2 gene in mouse ES cells suggesting that ncRNA might target polycomb repression at this specific locus. According to the “epigenetic switch” hypothesis, polycomb repression can be replaced by DNA methylation;Citation26 this could explain why polycomb target genes are predisposed to hypermethylation in cancers.Citation9-Citation11 Moreover, it has been reported that modifications in the DNA methylome can alter polycomb binding and repressive pattern.Citation27-Citation29 Future studies will need to clarify the interplay between DNMTs and polycomb proteins to understand which one triggers the upsetting of the other one and in which context.
In conclusion, the data presented here provide the first evidence that H3K27me3 and DNA methylation are related and co-exist in CpG rich regions in primary tissue samples from prostate cancer patients.
Materials and Methods
Prostate tumors
Patients’ samples were obtained after informed consent in accordance with the Declaration of Helsinki and stored at the « CRB Cancer des Hôpitaux de Toulouse » collection. According to the French law, CRB Cancer collection has been declared to the Ministry of Higher Education and Research (DC-2008-463) and obtained a transfer agreement (AC-2008-820) after approbation by ethical committees. Clinical and biological annotations of the samples have been declared to the CNIL (Comité National Informatique et Libertés). None of the six patients have received radiotherapy or hormonotherapy before prostate surgery. The six prostate tumors were freshly frozen after surgery to guarantee in vivo condition of analysis.
Bisulfite conversion followed by pyrosequencing
500 ng of genomic DNA was purified (DNeasy Blood and Tissue Kit, Qiagen) and bisulfite-converted (EZ DNA Methylation kit, Zymo Research) following the manufacturer's instructions. Quantitative DNA methylation analysis was performed by pyrosequencing on bisulfite treated DNA.Citation30 The region of interest validation was amplified using 30 ng of bisulfite treated human genomic DNA and 5 pmol of forward (5′-AGGAGGGTTT ATTTTTTGTT AAAGG) and reverse biotinylated primer (5′- Biotin –AAATTCTCCT TCCAAATAAA TACTTACAA). Reaction conditions were 1x HotStar Taq buffer supplemented with 1.6 mM MgCl2, 100 µM dNTPs and 2.0 U HotStar Taq polymerase (Qiagen) in a 25 µL volume. The PCR program consisted of a denaturing step of 15 min at 95 °C followed by 50 cycles of 30 s at 95 °C, 30 s at 61 °C and 20 s at 72 °C, with a final extension of 5 min at 72 °C. Ten microliters of PCR product were rendered single-stranded as previously describedCitation30 and 4 pmol of the respective sequencing primer (5′-TTGAGGATTG GGATGT for CpGs 1–5 and 5′-AGGGTTTGTT TGGGT for CpGs 6–10) were used for analysis. Quantitative DNA methylation analysis was performed on a PSQ 96MD system with the PyroGold SQA Reagent Kit and results were analyzed using the Q-CpG software (V.1.0.9, Pyrosequencing AB). For ChIP samples the immunoprecipitated DNA was bisulfite converted using the EpiTECT Plus kit (Qiagen) and the pyrosequencing was performed as previously described with the sole modification of using 3–4 µL of bisulfite converted DNA as input for PCR amplification. The average percentage of the methylation measured at the 10 CpGs located in the RARβ2 promoter is reported.
ChIP assays
Tumor samples were thawed on ice and fixed in 1% formaldehyde for 10 min at RT. The crosslinking reaction was stopped by incubation with 0.125 M of glycine for 5 min at RT. After centrifugation, tumors were resuspended in dissociation buffer (50 mM Hepes-KOH pH7.5, 140 mM NaCl, 1 mM EDTA, 10% glycerol, 0.5% NP-40, 0.25% Triton X100 and protease inhibitor) and fully dissociated with a glass dounce homogenizer. After centrifugation, cell pellets were incubated 10 min at 4 °C in a lysis buffer (200 mM NaCl, 1 mM EDTA, 0.5 mM EGTA, 10 mM Tris pH 8 and protease inhibitor). Then, tumors were sonicated for 20 min using cycles of 30 s ON – 30 s OFF (Diagenode Bioruptor). Sheared chromatin was pre-cleared and incubated at 4 °C overnight with 1 μg of the following primary antibodies: anti-H3K9me3 (Abcam), anti-H3K27me3 (Abcam), anti-RNAP II (Millipore) and non-specific IgG (Millipore). Chromatin-antibody complexes were precipitated with agarose beads and washed four times (Low salt buffer: 20 mM Tris pH8, 0.1% SDS, 1% Triton X100, 2 mM EDTA and 150 mM NaCl. High salt buffer: 20 mM Tris pH8, 0.1% SDS, 1% Triton X100, 2 mM EDTA and 500 mM NaCl. Li buffer: 0.25 mM LiCl, 1% NP-40, 1% Na-Deoxycholate, 10 mM Tris pH8 and 1 mM EDTA. TE buffer: 50 mM Tris pH8, 50 mM NaCl and 1mM EDTA). Eluted and de-crosslinked samples were treated with RNase A (0.3 µg/µL) and Proteinase K (0.2 µg/µL) and DNA was purified by phenol-chloroform extraction. The immunoprecipitated DNA and input samples were analyzed by real-time qPCR using SYBR Green (Applied Biosystems) according to the manufacturer's instructions. Primers used were as follow: RARβ2 5′-ATCCTGGGAG TTGGTGATGT CAG-3′ and 5′-AAAGAATAGA CCCTCCTGCC TC-3′, GAPDH 5′-TACTAGCGGT TTTACGGGCG-3′ and 5′-TCGAACAGGA GGAGCAGAGA GCGA-3′, MYT1 5′- CCAAGGGTTC ATGGGTAGCG TATT-3′ and 5′-GTGCGAACTC CTAAGCCAGC TAAA-3′.
| Abbreviations: | ||
| ChIP | = | chromatin immunoprecipitation |
| ChIP-BS-pyro | = | ChIP bisulfite pyrosequencing |
| ChIP-BS-seq | = | ChIP bisulfite sequencing |
| DNMT | = | DNA methyltransferase |
| ES cells | = | embryonic stem cells |
| EZH2 | = | enhancer of zeste homolog 2 |
| GAPDH | = | glyceraldehyde-3-phosphate dehydrogenase |
| H3K9me3 | = | trimethylated lysine 9 of histone H3 |
| H3K27me3 | = | trimethylated lysine 27 of histone H3 |
| MYT1 | = | myelin transcription factor 1 |
| ncRNA | = | non-coding RNA |
| PRC2 | = | polycomb repressive complex 2 |
| RA | = | retinoic acid |
| RARβ | = | retinoic acid receptor beta |
| RNAP II | = | RNA polymerase II |
Additional material
Download Zip (113.7 KB)Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
The authors acknowledge the Service d'Anatomie et Cytologie Pathologiques des Hôpitaux de Toulouse for the tumor samples. The authors thank Slimane Ait-Si-Ali and Julien Pontis (Université Paris Diderot) for their ChIP protocol and Loïc Ponger (MNHN, Paris) for critical discussions. This work was supported by ATIP CNRS and Equipe d’Excellence Région Midi-Pyrenées grants to PBA. CM was recipient of an MNERT fellowship.
References
- Kondo Y, Shen L, Cheng AS, Ahmed S, Boumber Y, Charo C, Yamochi T, Urano T, Furukawa K, Kwabi-Addo B, et al. Gene silencing in cancer by histone H3 lysine 27 trimethylation independent of promoter DNA methylation. Nat Genet 2008; 40:741 - 50; http://dx.doi.org/10.1038/ng.159; PMID: 18488029
- Lindroth AM, Park YJ, McLean CM, Dokshin GA, Persson JM, Herman H, Pasini D, Miró X, Donohoe ME, Lee JT, et al. Antagonism between DNA and H3K27 methylation at the imprinted Rasgrf1 locus. PLoS Genet 2008; 4:e1000145; http://dx.doi.org/10.1371/journal.pgen.1000145; PMID: 18670629
- Takeshima H, Yamashita S, Shimazu T, Niwa T, Ushijima T. The presence of RNA polymerase II, active or stalled, predicts epigenetic fate of promoter CpG islands. Genome Res 2009; 19:1974 - 82; http://dx.doi.org/10.1101/gr.093310.109; PMID: 19652013
- Brinkman AB, Gu H, Bartels SJ, Zhang Y, Matarese F, Simmer F, Marks H, Bock C, Gnirke A, Meissner A, et al. Sequential ChIP-bisulfite sequencing enables direct genome-scale investigation of chromatin and DNA methylation cross-talk. Genome Res 2012; 22:1128 - 38; http://dx.doi.org/10.1101/gr.133728.111; PMID: 22466170
- Statham AL, Robinson MD, Song JZ, Coolen MW, Stirzaker C, Clark SJ. Bisulfite sequencing of chromatin immunoprecipitated DNA (BisChIP-seq) directly informs methylation status of histone-modified DNA. Genome Res 2012; 22:1120 - 7; http://dx.doi.org/10.1101/gr.132076.111; PMID: 22466171
- Moison C, Senamaud-Beaufort C, Fourrière L, Champion C, Ceccaldi A, Lacomme S, Daunay A, Tost J, Arimondo PB, Guieysse-Peugeot AL. DNA methylation associated with polycomb repression in retinoic acid receptor β silencing. FASEB J 2013; 27:1468 - 78; http://dx.doi.org/10.1096/fj.12-210971; PMID: 23299856
- Houle B, Rochette-Egly C, Bradley WE. Tumor-suppressive effect of the retinoic acid receptor beta in human epidermoid lung cancer cells. Proc Natl Acad Sci U S A 1993; 90:985 - 9; http://dx.doi.org/10.1073/pnas.90.3.985; PMID: 8381540
- Seewaldt VL, Johnson BS, Parker MB, Collins SJ, Swisshelm K. Expression of retinoic acid receptor beta mediates retinoic acid-induced growth arrest and apoptosis in breast cancer cells. Cell Growth Differ 1995; 6:1077 - 88; PMID: 8519684
- Ohm JE, McGarvey KM, Yu X, Cheng L, Schuebel KE, Cope L, Mohammad HP, Chen W, Daniel VC, Yu W, et al. A stem cell-like chromatin pattern may predispose tumor suppressor genes to DNA hypermethylation and heritable silencing. Nat Genet 2007; 39:237 - 42; http://dx.doi.org/10.1038/ng1972; PMID: 17211412
- Schlesinger Y, Straussman R, Keshet I, Farkash S, Hecht M, Zimmerman J, Eden E, Yakhini Z, Ben-Shushan E, Reubinoff BE, et al. Polycomb-mediated methylation on Lys27 of histone H3 pre-marks genes for de novo methylation in cancer. Nat Genet 2007; 39:232 - 6; http://dx.doi.org/10.1038/ng1950; PMID: 17200670
- Widschwendter M, Fiegl H, Egle D, Mueller-Holzner E, Spizzo G, Marth C, Weisenberger DJ, Campan M, Young J, Jacobs I, et al. Epigenetic stem cell signature in cancer. Nat Genet 2007; 39:157 - 8; http://dx.doi.org/10.1038/ng1941; PMID: 17200673
- Viré E, Brenner C, Deplus R, Blanchon L, Fraga M, Didelot C, Morey L, Van Eynde A, Bernard D, Vanderwinden JM, et al. The Polycomb group protein EZH2 directly controls DNA methylation. Nature 2006; 439:871 - 4; http://dx.doi.org/10.1038/nature04431; PMID: 16357870
- Mohammad HP, Cai Y, McGarvey KM, Easwaran H, Van Neste L, Ohm JE, O’Hagan HM, Baylin SB. Polycomb CBX7 promotes initiation of heritable repression of genes frequently silenced with cancer-specific DNA hypermethylation. Cancer Res 2009; 69:6322 - 30; http://dx.doi.org/10.1158/0008-5472.CAN-09-0065; PMID: 19602592
- Cho NY, Kim BH, Choi M, Yoo EJ, Moon KC, Cho YM, Kim D, Kang GH. Hypermethylation of CpG island loci and hypomethylation of LINE-1 and Alu repeats in prostate adenocarcinoma and their relationship to clinicopathological features. J Pathol 2007; 211:269 - 77; http://dx.doi.org/10.1002/path.2106; PMID: 17139617
- Bovenzi V, Lê NL, Côte S, Sinnett D, Momparler LF, Momparler RL. DNA methylation of retinoic acid receptor beta in breast cancer and possible therapeutic role of 5-aza-2′-deoxycytidine. Anticancer Drugs 1999; 10:471 - 6; http://dx.doi.org/10.1097/00001813-199906000-00007; PMID: 10477167
- Sirchia SM, Ferguson AT, Sironi E, Subramanyan S, Orlandi R, Sukumar S, Sacchi N. Evidence of epigenetic changes affecting the chromatin state of the retinoic acid receptor beta2 promoter in breast cancer cells. Oncogene 2000; 19:1556 - 63; http://dx.doi.org/10.1038/sj.onc.1203456; PMID: 10734315
- Widschwendter M, Berger J, Hermann M, Müller HM, Amberger A, Zeschnigk M, Widschwendter A, Abendstein B, Zeimet AG, Daxenbichler G, et al. Methylation and silencing of the retinoic acid receptor-beta2 gene in breast cancer. J Natl Cancer Inst 2000; 92:826 - 32; http://dx.doi.org/10.1093/jnci/92.10.826; PMID: 10814678
- Antequera F, Boyes J, Bird A. High levels of de novo methylation and altered chromatin structure at CpG islands in cell lines. Cell 1990; 62:503 - 14; http://dx.doi.org/10.1016/0092-8674(90)90015-7; PMID: 1974172
- Ke XS, Qu Y, Cheng Y, Li WC, Rotter V, Øyan AM, Kalland KH. Global profiling of histone and DNA methylation reveals epigenetic-based regulation of gene expression during epithelial to mesenchymal transition in prostate cells. BMC Genomics 2010; 11:669; http://dx.doi.org/10.1186/1471-2164-11-669; PMID: 21108828
- Ke XS, Li WC, Hovland R, Qu Y, Liu RH, McCormack E, Thorsen F, Olsen JR, Molven A, Kogan-Sakin I, et al. Reprogramming of cell junction modules during stepwise epithelial to mesenchymal transition and accumulation of malignant features in vitro in a prostate cell model. Exp Cell Res 2011; 317:234 - 47; http://dx.doi.org/10.1016/j.yexcr.2010.10.009; PMID: 20969863
- Kirmizis A, Bartley SM, Kuzmichev A, Margueron R, Reinberg D, Green R, Farnham PJ. Silencing of human polycomb target genes is associated with methylation of histone H3 Lys 27. Genes Dev 2004; 18:1592 - 605; http://dx.doi.org/10.1101/gad.1200204; PMID: 15231737
- Bracken AP, Dietrich N, Pasini D, Hansen KH, Helin K. Genome-wide mapping of Polycomb target genes unravels their roles in cell fate transitions. Genes Dev 2006; 20:1123 - 36; http://dx.doi.org/10.1101/gad.381706; PMID: 16618801
- Varambally S, Dhanasekaran SM, Zhou M, Barrette TR, Kumar-Sinha C, Sanda MG, Ghosh D, Pienta KJ, Sewalt RG, Otte AP, et al. The polycomb group protein EZH2 is involved in progression of prostate cancer. Nature 2002; 419:624 - 9; http://dx.doi.org/10.1038/nature01075; PMID: 12374981
- Moison C, Arimondo PB, Guieysse-Peugeot AL. Commercial reverse transcriptase as source of false-positive strand-specific RNA detection in human cells. Biochimie 2011; 93:1731 - 7; http://dx.doi.org/10.1016/j.biochi.2011.06.005; PMID: 21689721
- Zhao J, Ohsumi TK, Kung JT, Ogawa Y, Grau DJ, Sarma K, Song JJ, Kingston RE, Borowsky M, Lee JT. Genome-wide identification of polycomb-associated RNAs by RIP-seq. Mol Cell 2010; 40:939 - 53; http://dx.doi.org/10.1016/j.molcel.2010.12.011; PMID: 21172659
- Gal-Yam EN, Egger G, Iniguez L, Holster H, Einarsson S, Zhang X, Lin JC, Liang G, Jones PA, Tanay A. Frequent switching of Polycomb repressive marks and DNA hypermethylation in the PC3 prostate cancer cell line. Proc Natl Acad Sci U S A 2008; 105:12979 - 84; http://dx.doi.org/10.1073/pnas.0806437105; PMID: 18753622
- Wu H, Coskun V, Tao J, Xie W, Ge W, Yoshikawa K, Li E, Zhang Y, Sun YE. Dnmt3a-dependent nonpromoter DNA methylation facilitates transcription of neurogenic genes. Science 2010; 329:444 - 8; http://dx.doi.org/10.1126/science.1190485; PMID: 20651149
- Reddington JP, Perricone SM, Nestor CE, Reichmann J, Youngson NA, Suzuki M, Reinhardt D, Dunican DS, Prendergast JG, Mjoseng H, et al. Redistribution of H3K27me3 upon DNA hypomethylation results in de-repression of Polycomb target genes. Genome Biol 2013; 14:R25; http://dx.doi.org/10.1186/gb-2013-14-3-r25; PMID: 23531360
- Reddington JP, Sproul D, Meehan RR. DNA methylation reprogramming in cancer: Does it act by re-configuring the binding landscape of Polycomb repressive complexes?. Bioessays 2014; 36:134 - 40; http://dx.doi.org/10.1002/bies.201300130; PMID: 24277643
- Tost J, Gut IG. DNA methylation analysis by pyrosequencing. Nat Protoc 2007; 2:2265 - 75; http://dx.doi.org/10.1038/nprot.2007.314; PMID: 17853883
