Abstract
Several recent publications demonstrate a co-activator function for a subgroup of plant homeodomain fingers, which in humans comprises PHF2, PHF8 and KIAA1718. Besides an N-terminal plant homeodomain (PHD) these proteins also harbor an enzymatically active Jumonji-C domain (JmjC). While they have been shown to bind via their PHDs to H3K4me3-bearing nucleosomes at active gene promoters, their JmjC-domains are able to remove mono- and dimethyl-lysine 9 or 27 on histone H3, and monomethyl-lysine 20 on histone H4, chromatin modifications which correlate with transcriptional repression. Such dual histone crosstalk insures the proper removal of repressive histone marks following transcriptional activation by RNA polymerases I and II. Mutations in the PHF8 gene lead to X-linked mental retardation (XLMR) and knockdown of KIAA1718 and PHF8 homologs in zebrafish causes brain defects. Thus, the co-activator function of this new class of chromatin modifying enzymes has important functional roles in neuronal development. To continue with the nomenclature for histone demethylases, we propose the usage of KDM7A, -B and -C for KIAA1718, PHF8 and PHF2 proteins.
Histone Modifications and Chromatin-Modifying Enzymes
A host of transcriptional co-activators and co-repressors function through post-translational modifications (PTMs) of histones.Citation1 An important and well-studied type of PTM in this respect is methylation of the e-aminogroup of the lysine sidechain, which can occur in three possible states, namely mono-, di- or trimethylation (me1/2/3, respectively). Depending on the histone variant, the position of the lysine and its methylation state, such modifications may signal different outcomes. For example, trimethylation at lysines 4, 36 and 79 of histone H3 (H3K4me3, H3K36me3, H3K79me3) are positively correlated with transcription and thus considered as activating chromatin marks. While H3K4me3 is strongly enriched at proximal promoters, H3K36me3 and H3K79me3 are found throughout transcribed gene bodies.Citation2 Conversely, di- and trimethylation at lysines 9 and 27 of histone H3 (H3K9me3/2, H3K27me3/2) spread on silent chromatin and therefore are often correlated with transcriptional repression.
Methyl-marks are deposited on lysine residues by protein complexes that contain lysine methyl transferase enzymes (KMTs), most of which contain a Su(var)3-9/Enhancer of Zeste/Trithorax (SET) domain. For instance, in humans H3K4-methylation is conferred by enzymes of the KMT2-family (SET1A/B and mixed lineage leukemia MLL1-5).Citation3 The latter are thought to be recruited by transcription factors upon gene induction and to methylate nucleosomes at the respective promoters. Subsequently, a diverse number of proteins recognizing H3K4me3 come into play, some of which have already been shown to stimulate transcription (e.g., TAF3, BPTF).Citation4,Citation5
On the other hand, methyl-groups can be removed from lysines by histone demethylase enzymes (KDMs) containing either flavin-dependent amine-oxidase or Fe2+ and α-ketoglutarate-dependent Jumonji-C (JmjC) domains.Citation6 The aforementioned methylation of lysine 4 on histone H3 can be removed by either LSD1/KDM1- or JARID1/KDM5-family histone demethylases.Citation3 Intriguingly, these KDMs can be components of polycomb repressive complexes (PRCs) that synergize to bring about transcriptional repression.Citation7,Citation8 Interestingly, in addition to their catalytic domains most chromatin-modifying enzymes possess additional chromatin and/or DNA-binding modules.Citation9 Such modular nature of chromatin-modifying enzymes allows for additional specificity in their recruitment and/or mode of action.Citation10 Therefore, it is likely that recruitment of such enzymes to their target is accomplished by multiple synergistic interactions of the components of the chromatin-modifying complex and different modifications of the chromatin. Such a mechanism would allow regulation of specificity as well as affinity by tissue-specific variation of subunit composition combined with dynamic changes in histone post-transcriptional modifications allowing for fine-tuning of gene expression in a tissue- and time-specific manner.
The Plant Homeodomain (PHD): A Chromatin Reader Module
Recently, we and others found a new example of crosstalk between histone modifications.Citation11–Citation17 A class of JmjC-domain chromatin-modifying enzymes is characterized by a single N-terminal plant homeodomain zinc finger (PHD), a domain that was shown to associate with methylated lysine residues.Citation18 In humans this group consists of three members: the plant homeodomain fingers 2 and 8 (PHF2 and PHF8) and KIAA1718 (). Structures of their PHDs and JmjC-domains have now been solved by X-ray crystallography.Citation13,Citation15,Citation19–Citation21 Besides conserved zinc-chelating residues, these PHDs comprise a patch of phenylalanine and tyrosine residues called aromatic cage, which can occur in several domains and can interact with methylated lysine. Biochemical experiments demonstrated that the three proteins interact specifically with histone H3 methylated at lysine 4 via their PHDs.Citation11–Citation15,Citation22 Mutation of aromatic cage amino acids abolishes this interaction. Binding assays with doubly modified histone H3 peptides indicate that most adjacent PTMs do not affect the interaction between H3K4me3 and the PHD of PHF8/KDM7B, but that phosphorylation of threonine 3 does, a modification mediated by the mitotic kinase haspin.Citation12,Citation23 Additionally, phosphorylation of PHF8 at serines 33 and 84 abolishes its binding to mitotic chromosomes.Citation16 However, during interphase, PHF8 is present at thousands of gene promoters in several cell lines as revealed by chromatin immunoprecipitation followed by deep sequencing (ChIP-seq) using different cell lines and antibodies.Citation12,Citation14,Citation16,Citation17 Similar results were obtained for F29B9.2/KDM7A in C. elegans.Citation24 Thus, we assume that the localization is mainly determined by binding of the PHD to H3K4-trimethylated nucleosomes, and that PHF2/KDM7C has similar properties. Nevertheless, about 30% of highly H3K4me3-enriched promoters are not co-occupied by PHF8, which indicates that there must be other factors influencing the recruitment.Citation12,Citation14,Citation16
Furthermore, there are exceptions to the rule that H3K4me3 is generally linked to active gene expression. For example, a class of PHD-containing proteins, the inhibitor of growth family (ING1-5), has been demonstrated to bind to H3K4me3 but to repress transcription upon DNA-damage.Citation25 On the other hand, the PHD-protein AIRE was shown to interact with unmethylated lysine 4 on histone H3, leading to transcriptional activation.Citation26 Additionally, there is a class of poised chromatin regions in mouse embryonic stem cells known as bivalent domains, which harbor both H3K4me3 and H3K27me3 chromatin modifications. In embryonic stem (ES) cells many developmental genes bear bivalent domains and are thought to be rapidly induced by removal of such repressive chromatin modifications upon differentiation.Citation27 This may be accomplished by displacement of PRCs with activating complexes that exhibit demethylase activity towards repressive marks.
The Jumonji C-Terminal Domain (JmjC) Confers Demethylation
Active JmjC-domains comprise several conserved amino acids that are needed for correct binding of the crucial cofactors Fe2+ and α-ketoglutarate (JmjC-containing KDM-families are listed in ). Besides those, the KDM7-family displays four short additional α-helices in the carboxyterminal region of the JmjC which are essential for activity. The described enzymes act on methylated H3K9, H3K27 and/or H4K20. Interestingly, H3K9 and H3K27 are placed in a similar context (-ARKS-), while H4K20 is not (-HRKV-). Activity has been demonstrated by in vitro assays using peptides, bulk histones or nucleosomes as substrates and in vivo by overexpression and/or knockdown experiments.Citation11–Citation17,Citation19,Citation20,Citation22,Citation28,Citation29 While demethylation assays with peptides also stated marginal activity towards H3K36me2, there is little evidence that these enzymes remove this histone modification in vivo. The three new members exhibit slightly different specificities (). While PHF8 preferentially acts on H3K9me2/1 and H4K20me1, KIAA1718 mainly demethylates H3K9me2/1 and H3K27me2/1.Citation13,Citation16,Citation17,Citation22,Citation28 Since the active centers cannot accommodate trimethylated lysines these do not constitute viable substrates for these enzymes. Intriguingly, JmjCs that exhibit aspartate as Fe2+-chelating residue seem to be specific for me2/1, while glutamate at this position confers specificity for me3/2 (). In contrast, thus far PHF2 was only shown to demethylate H3K9me1.Citation15 The difference might be due to the fact that this protein, instead of the second conserved Fe2+-binding histidine, contains a tyrosine residue at this position. Trimethylation of H3K4 in cis greatly enhances PHF8 demethylase activity toward H3K9me2.Citation11,Citation13 However, while PHF8 can mediate H3K4me3-binding through its PHD domain and H3K9me2-demethylation on the same histone tail as a result of its flexible linker between PHD and JmjC, KIAA1718 has a rigid linker and an extended N-terminus, precluding it from demethylation of H3K9me2 once its PHD associates with H3K4me3.Citation13 When H3K4me3 peptides are added to H3K9me2-demethylation reactions in trans KIAA1718 is inhibited and PHF8 is not stimulated anymore.Citation11,Citation13 Nevertheless, in vivo H3K4me3 might as well stimulate demethylation of H3K9me2/1, H3K27me2/1 and H4K20me1 on neighboring histone tails because it anchors the KDM7 enzymes in close proximity.Citation21 This possibility may explain why H4K20me1 demethylation could only be detected on nucleosomal substrates but not on core histones.Citation16,Citation17 Obviously, H3K4me3-marks and a functional PHD stimulate demethylation by overexpression of PHF8 in vivo.Citation12 Consequently, chromatin-association of PHF8 via the interaction between PHD and H3K4me3 is crucial for proper function. On the other hand, though mutations of cofactor binding residues in the JmjC render PHF8 catalytically inactive, it still retains most of its co-activation capacity, at least in episomal reporter assays.Citation12 Thus, we expect that additional protein interactions are of particular importance for proper function of KDM7 family members.
The Carboxyterminal Half and Interaction Partners
Little is known about the properties of the C-terminal halves of KDM7 proteins. They do not contain any known protein domains, and the homology between the three human proteins is low in this region compared to that of PHD and JmjC (). However, we found that direct association of PHF8 with the carboxyterminal domain of RNA polymerase II (RNAPII) largest subunit strongly depends on this part of the protein.Citation12 Other described interactions of PHF8 with RNA polymerase I (RNAPI), KMT2-complex components, HCF-1, E2F1, ZNF711 and RAR may also be mediated by its C-terminal part ().Citation11,Citation14,Citation16,Citation30 The KDM7 C-termini contain nuclear localization signals suggesting their importance for correct localization (). Additionally, all three human proteins contain a putative coiled coil region (). PHF2 and PHF8 also exhibit several phosphorylation sites which appear to be important for the regulation of their activity ().Citation16
Biological Function and Target Genes
Recent findings point to the KDM7-family of histone demethylases as a new class of transcriptional co-activators. By virtue of association with H3K4me3- and removal of H3K9me2/1-, H3K27me2/1-or H4K20me1-modifications, KDM7 proteins create a more permissive chromatin environment at promoters (). Only then the respective demethylated lysine residues can be targeted for acetylation by acetyltransferases, which may lead to enhanced expression. Moreover, additional interactions with RNAPI, RNAPII and other proteins may further stimulate transcription and could play a role in the regulation of activity. Furthermore, other unknown (non-)histone proteins could be substrate for binding and/or demethylation by KDM7-members and nascent RNA could be involved in this process as well.Citation31
Importantly, mutations in the PHF8 gene can cause Siderius-Hamel syndrome, an X-linked mental retardation (XLMR), which is often accompanied by cleft lip and/or cleft palate.Citation32 The mutations mostly lead to truncations before or in the JmjC domain; however, a complete deletion of the locus and a point mutation has also been described.Citation33–Citation36 These aberrations cause loss of function as demonstrated for the XLMR point mutant F279S, which mislocalizes probably due to misfolding of the JmjC domain. Depletion of PHF8 does not result in a global increase of histone methylation but in a rather modest increase of H4K20me1 and H3K9me1-levels at target promoters.Citation16,Citation17 Surprisingly, H3K9me2, H3K27me2 and H3K36me2 remain mostly unchanged. In contrast, loss of one of the two homologs in C. elegans (F29B9.2) leads to global increase in H3K9me2 as well as H3K27me2 and affects locomotion.Citation14 In addition, RNA interference-mediated depletion of PHF8-or KIAA1718-homologs disturbs neuronal differentiation of mouse ES-cells and causes defects in brain and craniofacial development in zebrafish.Citation17,Citation22,Citation28,Citation30 Liu et al. suggests that the loss of PHF8 affects cell cycle progression due to impaired co-activation of E2F1 targets.Citation16 However, the specific defects of XLMR patients in neuronal and midline development may not be a reflection of the general function of PHF8 in growth but rather result from a specific role PHF8 may play in neuronal proliferation.Citation17 PHF8 function could be critical for neurons as its activity may not be compensated by other family members in neuronal cells or that it may have neuronal-specific targets.
PHF8 and PHF2 both play a role as co-activators of ribosomal RNA transcription where they might act redundantly.Citation11,Citation15,Citation37 Though knockdown of PHF8 results in downregulation of several RNAPII-transcribed genes, changes in histone methylation and expression levels are subtle and there is little overlap between targets found in different studies.Citation12,Citation14,Citation16,Citation17 Interestingly, one candidate target gene, SMCX (also known as JARID1C or KDM5C) codes for another KDM linked to XLMR, which also contains PHDs and a JmjC domain. In contrast to KDM7, KDM5C demethylates H3K4me3/2, binds to H3K9me3 and acts as a transcriptional repressor.Citation8 Overexpression of other candidate targets like FGF4, FST and MSX1 can in part rescue the loss of KDM7 proteins in model systems.Citation17,Citation22,Citation28
Taken together, it is not clear whether the PHF8 mutations leading to impairment of transcription by RNAPI, RNAPII or both accounts for manifestation of XLMR. The gene expression changes are rather subtle and therefore it is possible that a combined decrease in transcriptional output of multiple targets may be the underlying cause of the disease phenotype. Further experiments using model organisms or XLMR-patients' material will shed additional insights on the target genes and biological functions of the KDM7-family members.
Abbreviations
| PHD | = | plant homeodomain |
| PHF2/8 | = | plant homeodomain finger 2/8 |
| JmjC | = | jumonji carboxyterminal domain |
| XLMR | = | X-(chromosome) linked mental retardation |
| PTM | = | post-translational modification |
| KMT | = | histone lysine methyl-transferase |
| SET | = | Su(var)3–9/enhancer of zeste/trithorax domain |
| MLL | = | mixed lineage leukemia |
| KDM | = | histone lysine demethylase |
| PRC | = | polycomb repressive complex |
| ChIP-seq | = | chromatin immunoprecipitation followed by deep sequencing |
| ING | = | inhibitor of growth |
| ES-cells | = | embryonic stem cells |
| RNAPI/II | = | RNA polymerase I/II |
Figures and Tables
Figure 1 KDM7-family proteins. In humans the KDM7-family comprises three members: KIAA1718, PHF8 and PHF2. These proteins of about 1,000 amino acids (a.a.) contain an N-terminal plant homeodomain (PHD), a Jumonji C-domain (JmjC) and a short coiled coil region (cc). Nuclear localization signals and phosphorylation sites are depicted as arrows below and arrowheads above the model, respectively. Homology between the family members is given in percent identity for PHD, JmjC-domain and C-terminal region defined by dashed lines.
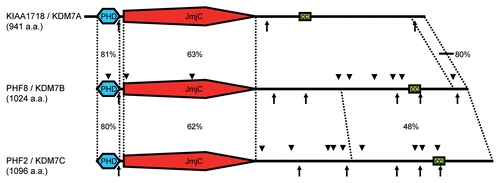
Figure 2 Simplified model of action for transcriptional co-activation by KDM7-proteins. Upon gene induction, transcription factors (TF) bind to specific DNA-sequences and recruit co-activators such as H3K4-methyltransferases (KMT2). The latter introduce methyl-marks on histone H3 tails (me-K4) which protrude from nucleosomes. These activation chromatin marks are bound by proteins such as histone demethylases of the KDM7-family. This class of chromatin-modifying enzymes removes repressive methyl-marks from K9 or K27 on H3 or K20 on H4. Furthermore KDM7 can interact with RNA polymerases I or II (RNAP) and other chromatin-associated factors (such as E2F1 and ZNF711). Both mechanisms lead to the activation of transcription. However, the transcription factors and interactions may be specific for cell type, time point and gene locus, which could be the means by which KDM7-activity is regulated.
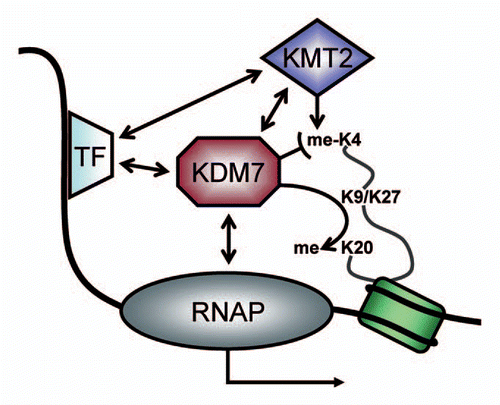
Table 1 Human KDM-families with JmjC-domains
Acknowledgements
K.F. received an Erwin-Schrödinger fellowship from the Austrian science fund FWF (J2728-B12). R.S. was supported by a grant from NIH (CA090758).
References
- Kouzarides T. Chromatin modifications and their function. Cell 2007; 128:693 - 705
- Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, et al. High-resolution profiling of histone methylations in the human genome. Cell 2007; 129:823 - 837
- Allis CD, Berger SL, Cote J, Dent S, Jenuwein T, Kouzarides T, et al. New nomenclature for chromatin-modifying enzymes. Cell 2007; 131:633 - 636
- Vermeulen M, Mulder KW, Denissov S, Pijnappel WW, van Schaik FM, Varier RA, et al. Selective anchoring of TFIID to nucleosomes by trimethylation of histone H3 lysine 4. Cell 2007; 131:58 - 69
- Wysocka J, Swigut T, Xiao H, Milne TA, Kwon SY, Landry J, et al. A PHD finger of NURF couples histone H3 lysine 4 trimethylation with chromatin remodelling. Nature 2006; 442:86 - 90
- Klose RJ, Kallin EM, Zhang Y. JmjC-domain-containing proteins and histone demethylation. Nat Rev Genet 2006; 7:715 - 727
- Lee MG, Norman J, Shilatifard A, Shiekhattar R. Physical and functional association of a trimethyl H3K4 demethylase and Ring6a/MBLR, a polycomb-like protein. Cell 2007; 128:877 - 887
- Tahiliani M, Mei P, Fang R, Leonor T, Rutenberg M, Shimizu F, et al. The histone H3K4 demethylase SMCX links REST target genes to X-linked mental retardation. Nature 2007; 447:601 - 605
- Taverna SD, Li H, Ruthenburg AJ, Allis CD, Patel DJ. How chromatin-binding modules interpret histone modifications: lessons from professional pocket pickers. Nat Struct Mol Biol 2007; 14:1025 - 1040
- Suganuma T, Workman JL. Crosstalk among histone modifications. Cell 2008; 135:604 - 607
- Feng W, Yonezawa M, Ye J, Jenuwein T, Grummt I. PHF8 activates transcription of rRNA genes through H3K4me3 binding and H3K9me1/2 demethylation. Nat Struct Mol Biol 2010; 17:445 - 450
- Fortschegger K, de Graaf P, Outchkourov NS, van Schaik FMA, Timmers HTM, Shiekhattar R. PHF8 targets histone methylation and RNA polymerase II to activate transcription. Mol Cell Biol 2010; 30:3286 - 3298
- Horton JR, Upadhyay AK, Qi HH, Zhang X, Shi Y, Cheng X. Enzymatic and structural insights for substrate specificity of a family of jumonji histone lysine demethylases. Nat Struct Mol Biol 2010; 17:38 - 43
- Kleine-Kohlbrecher D, Christensen J, Vandamme J, Abarrategui I, Bak M, Tommerup N, et al. A functional link between the histone demethylase PHF8 and the transcription factor ZNF711 in X-linked mental retardation. Mol Cell 2010; 38:165 - 168
- Wen H, Li J, Song T, Lu M, Kan PY, Lee MG, et al. Recognition of histone H3K4 trimethylation by the plant homeodomain of PHF2 modulates histone demethylation. J Biol Chem 2010; 285:9322 - 9326
- Liu W, Tanasa B, Tyurina OV, Zhou TY, Gassmann R, Liu WT, et al. PHF8 mediates histone H4 lysine 20 demethylation events involved in cell cycle progression. Nature 2010; 466:508 - 512
- Qi HH, Sarkissian M, Hu GQ, Wang Z, Bhattacharjee A, Gordon DB, et al. Histone H4K20/H3K9 demethylase PHF8 regulates zebrafish brain and craniofacial development. Nature 2010; 466:503 - 507
- Li H, Ilin S, Wang W, Duncan EM, Wysocka J, Allis CD, et al. Molecular basis for site-specific read-out of histone H3K4me3 by the BPTF PHD finger of NURF. Nature 2006; 442:91 - 95
- Yu L, Wang Y, Huang S, Wang J, Deng Z, Zhang Q, et al. Structural insights into a novel histone demethylase PHF8. Cell Res 2010; 20:166 - 173
- Yue WW, Hozjan V, Ge W, Loenarz C, Cooper CD, Schofield CJ, et al. Crystal structure of the PHF8 Jumonji domain, an Nepsilon-methyl lysine demethylase. FEBS Lett 2010; 584:825 - 830
- Yang Y, Hu L, Wang P, Hou H, Lin Y, Liu Y, et al. Structural insights into a dual-specificity histone demethylase ceKDM7A from Caenorhabditis elegans. Cell Res 2010; 20:886 - 898
- Tsukada Y, Ishitani T, Nakayama KI. KDM7 is a dual demethylase for histone H3 Lys 9 and Lys 27 and functions in brain development. Genes Dev 2010; 24:432 - 437
- Dai J, Sultan S, Taylor SS, Higgins JM. The kinase haspin is required for mitotic histone H3 Thr 3 phosphorylation and normal metaphase chromosome alignment. Genes Dev 2005; 19:472 - 488
- Lin H, Wang Y, Tian F, Pu P, Yu Y, Mao H, et al. Coordinated regulation of active and repressive histone methylations by a dual-specificity histone demethylase ceKDM7A from Caenorhabditis elegans. Cell Res 2010; 20:899 - 907
- Soliman MA, Riabowol K. After a decade of study-ING, a PHD for a versatile family of proteins. Trends Biochem Sci 2007; 32:509 - 519
- Org T, Rebane A, Kisand K, Laan M, Haljasorg U, Andreson R, et al. AIRE activated tissue specific genes have histone modifications associated with inactive chromatin. Hum Mol Genet 2009; 18:4699 - 4710
- Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell 2006; 125:315 - 326
- Huang C, Xiang Y, Wang Y, Li X, Xu L, Zhu Z, et al. Dual-specificity histone demethylase KIAA1718 (KDM7A) regulates neural differentiation through FGF4. Cell Res 2010; 20:154 - 165
- Loenarz C, Ge W, Coleman ML, Rose NR, Cooper CD, Klose RJ, et al. PHF8, a gene associated with cleft lip/palate and mental retardation, encodes for an Nepsilon-dimethyl lysine demethylase. Hum Mol Genet 2010; 19:217 - 222
- Qiu J, Shi G, Jia Y, Li J, Wu M, Dong S, et al. The X-linked mental retardation gene PHF8 is a histone demethylase involved in neuronal differentiation. Cell Res 2010; 20:908 - 918
- Core LJ, Waterfall JJ, Lis JT. Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science 2008; 322:1845 - 1848
- Siderius LE, Hamel BC, van Bokhoven H, de Jager F, van den Helm B, Kremer H, et al. X-linked mental retardation associated with cleft lip/palate maps to Xp11.3-q21.3. Am J Med Genet 1999; 85:216 - 220
- Abidi FE, Miano MG, Murray JC, Schwartz CE. A novel mutation in the PHF8 gene is associated with X-linked mental retardation with cleft lip/cleft palate. Clin Genet 2007; 72:19 - 22
- Koivisto AM, Ala-Mello S, Lemmela S, Komu HA, Rautio J, Jarvela I. Screening of mutations in the PHF8 gene and identification of a novel mutation in a Finnish family with XLMR and cleft lip/cleft palate. Clin Genet 2007; 72:145 - 149
- Laumonnier F, Holbert S, Ronce N, Faravelli F, Lenzner S, Schwartz CE, et al. Mutations in PHF8 are associated with X linked mental retardation and cleft lip/cleft palate. J Med Genet 2005; 42:780 - 786
- Qiao Y, Liu X, Harvard C, Hildebrand MJ, Rajcan-Separovic E, Holden JJ, et al. Autism-associated familial microdeletion of Xp11.22. Clin Genet 2008; 74:134 - 144
- Zhu Z, Wang Y, Li X, Xu L, Wang X, Sun T, et al. PHF8 is a histone H3K9me2 demethylase regulating rRNA synthesis. Cell Res 2010; 20:794 - 801