Abstract
Testicular germ cell tumors are the most frequent malignant tumors in young Caucasian males, with increasing incidence. The actual model of tumorigenesis is based on the theory that a block in maturation of fetal germ cells lead to formation of the intratubular germ cell neoplasia unclassified. Early fetal germ cells and undifferentiated germ cell tumors express pluripotency markers such as the transcription factor NANOG. It has been demonstrated, that epigenetic modifications such as promoter DNA-methylation is able to silence gene expression in normal and cancer cells. Here we show, that OCT3/4-SOX2 mediated expression of NANOG can be silenced by methylation of promoter CpG-sites. We found that global methylation of DNA decreased from fetal spermatogonia to mature sperm. In contrast, CpGs in the NANOG promoter were found hypomethylated in spermatogonia and hypermethylated in sperm. This selective repression might reflect the cells need to suppress pluripotency in order to prevent malignant transformation. Finally, methylation of CpGs in the NANOG promoter in germ cell tumors and derived cell lines correlated to differentiation state.
Introduction
Methylation of cytosine residues within the genomic sequence of CpG islands and/or promoter regions is known to result in regulation of transcriptional activity during embryogenesis and differentiation. This epigenetic modification is crucial for developmental processes, including genomic imprinting and silencing of promoters within the human genome.Citation1 CpGs are overall underrepresented in the mammalian genome, while short CpG-rich regions with a CpG-density of >60%, called CpG islands, are found in promoter regions of almost 50% of all genes. These regions are usually hypomethylated in normal cells, with the exception of imprinted genes, the latter in a parent-dependent pattern.Citation1
NANOG is a key regulator of self-renewal and maintenance of pluripotency in undifferentiated embryonic stem cells.Citation2,Citation3 NANOG is expressed in the inner cell mass (ICM) of the blastocyst, as well as the epiblast at post implantation stage and is detectable in germ cells, seminoma, embryonal carcinoma and carcinoma in situ (CIS), also referred to as Intratubular Germ Cell Neoplasia Unclassified (IGCNU).Citation3–Citation5 NANOG expression is not detectable in the adult testis or in differentiated somatic cells.Citation6 The protein contains a DNA-binding domain, which is important for transcriptional regulation of developmental key processes in combination with other proteins like OCT3/4 and SOX2. Mitsui and Chambers demonstrated that overexpression of NANOG enables embryonic stem cells (ESCs) to keep up self-renewing abilities independent of the LIF/STAT-pathway.Citation3,Citation4 Deletion of NANOG triggers ESCs to differentiate into parietal/visceral endoderm, revealing its role in the second embryonic differentiation event.Citation7,Citation8 These data underline NANOG's important role in maintenance of pluripotency and in suppression of differentiation.
During mammalian embryogenesis primordial germ cells (PGCs) are specified by BMP-signals (BMP4/BMP8b).Citation9 These cells migrate along the hindgut to the genital ridges, which develop to the gonads. During their migration PGCs express pluripotency markers, like NANOG and OCT3/4. At the genital ridges PGCs differentiate into fetal spermatogonia, which settle down at the basal membranes of the seminiferous tubules and maturate into sperm during spermatogenesis. Expression of NANOG and OCT3/4 becomes downregulated upon transition to fetal spermatogonia.Citation10
Germ cell tumors (GCTs) consist of a heterogeneous group, which is classified into five subtypes according on their different biological characteristics and their origin.Citation11 Malignant seminomatous and non-seminomatous GCTs occur most frequently in the testicles.Citation12 Seminomas are undifferentiated cells that lack SOX2, but express SOX17 instead.Citation13 The non-seminomas can further be divided into subgroups: (1) the undifferentiated, pluripotent embryonal carcinomas (EC), which are able to differentiate into (2) more differentiated tumors including teratoma, yolk sac tumor and choriocarcinoma.
Here, we show that human NANOG expression is mediated by a promoter element in the 5′ region upstream exon 1 (NANOG regulatory region; NRR) of the NANOG locus and depending on transactivation by OCT3/4 and SOX2, as well as on NRR DNA methylation. We demonstrate that a lack of NANOG expression in fetal spermatogonia is not due to epigenetic repression, but rather a result of lack of transcriptional activators such as OCT3/4 and SOX2. Our findings further suggest that epigenetic silencing of NANOG expression during germ cell maturation is established at post-spermatogonial state and is independent of global DNA methylation. We show, that the methylation profile of the active NRR in GCTs and corresponding cell lines correlates with NANOG expression detected by qRT-PCR and western blot and with the differentiation state of the germ cell tumor entity. Hence, the analysis of the NRR DNA methylation profile may serve as a diagnostic tool for human GCTs and GCT-derived cell lines.
Results
First, we sought to identify a sequence of the NANOG promoter, which could be the target of epigenetic modification and might be involved in regulation of expression. The region upstream of the transcriptional start site (TSS) of the human NANOG gene has been described to contain functional binding sites for OCT3/4 and SOX2Citation17–Citation19 at −180 bp and shown to be hypermethylated in human NT2 cells during neuronal differentiation.Citation20 It will be referred to as NANOG regulatory region (NRR; spanning bp −306-bp-1, Sup. Fig. 1A). Using PromoterScan1.7 software (www-bimas.cit.nih.gov/molbio/proscan/) we were able to verify this region (Sup. Fig. 1B, first yellow box). The element containing the TATA-box and the transcription start site encodes for CpG-dinucleotides, which could be the target of epigenetic modification. A sequence comparison of murine, cow, chimp and human Nanog/NANOG promoters revealed that the CpG-dinucleotides are conserved completely in chimp and human (Sup. Fig. 1B) suggesting that DNA methylation and underlying mechanisms might be conserved between those closely related species.
Next, we addressed the question of whether OCT3/4 and SOX2 are able to transactivate the NRR and if the transactivation could be silenced by methylation of CpG-sites within this sequence. Therefore, the NRR was cloned 5′ of a luciferase gene and transfected into the embryonal carcinoma cell line 2102EP, which expresses both OCT3/4 and SOX2.Citation15 Luciferase activity was strongly induced by the NRR compared to the empty reporter construct (). Transfection of a NRR harboring mutated OCT3/4-SOX2 binding sites or transfection of an in vitro methylated NRR resulted in a significantly reduced luciferase activity (p = 0.002; p = 0.001, respectively). Additionally, transfection of an in vitro methylated NRR harboring mutated OCT3/4-SOX2 binding sites resulted in a complete loss of transactivation. To further analyze the influence of NRR-methylation and expression of NANOG, the demethylating drug 5-aza-2′-deoxycytidine (5-azaC) was used to treat JKT-1 cells, which show a high level of methylation.Citation1 Treatment with 5-azaC led to a decrease in methylation at the NRR () and induction of NANOG expression (). These results suggested that the OCT3/4-SOX2 sites are of central importance for OCT3/4-SOX2 mediated NANOG transactivation and that this transactivation is silenced by methylation of the five CpGs within the NRR that are in close vicinity to the binding sites of OCT3/4 and SOX2 (Sup. Fig. 1).
Next, we addressed the question whether NANOG expression correlates to NRR CpG-methylation in human germ cells during development. NANOG is expressed in fetal gonocytes, becomes downregulated in fetal spermatogoniaCitation5 and is absent in postnatal gonads.Citation6 To analyze the methylation of the NRR during human male germ cell development, fetal gonocytes (19th week of gestation) and fetal spermatogonia (24th week of gestation) were detected using c-KIT and MAGE-A4 immunohistochemistry respectivelyCitation21 and collected by laser-capture-microdissection ( and B). Interestingly, the NRR is found to be completely demethylated in fetal gonocytes (), fetal spermatogonia () and spermatogonia of adults irrespective of age (). The CpGs in human adult testis tissue () and sperm () revealed hypermethylation of the NRR. This suggests that the CpGs of the NRR become hypermethylated during maturation of spermatogonia to sperm and that lack of NANOG expression at the fetal spermatogonia and spermatogonia stage is not due to methylation-mediated silencing at the NRR.
Next, we examined the expression of the NANOG transactivators OCT3/4 and SOX2 in fetal gonocytes (week 19 of gestation) and fetal spermatogonia (week 24 of gestation). In fetal germ cells at week 19 of gestation only OCT3/4, but not SOX2 could be detected ( and C). In fetal spermatogonia (24th week of gestation) neither OCT3/4 nor SOX2 expression were detected ( and D). So, NANOG expression in fetal gonocytes might be transactivated by OCT3/4 and SOX17, which compensates for the lack of SOX2.Citation13 In fetal spermatogonia the lack of NANOG expression can be explained by the lack of expression of its transactivators OCT3/4 and SOX2.
In order to compare global DNA methylation to methylation of CpGs at the NRR immunohistochemical staining of testis tissues from patients of age 2–29 years was performed. DNA of spermatogonia was found globally hypermethylated (, red arrows) while DNA of round spermatids was hypomethylated ( and D, black arrowheads). This result is in contrast to the specific demethylation of the CpGs at the NRR in spermatogonia (compare to ) and the hypermethylation in sperm (). So, the specific NRR DNA methylation profiles of spermatogonia and sperm did not follow the status of the global DNA methylation.
Next, we determined the methylation of CpGs in the NRR and the expression of NANOG in various GCTs. In seminomas and ECs, both being positive for NANOG protein, the CpGs of the NRR were hypomethylated ( and B).Citation6,Citation22 Teratomas, yolk sac tumors and choriocarcinomas representing NANOG-negative GCTs showed strong methylation of the NRR ().Citation6,Citation22 Mixed non-seminomas, containing undifferentiated and differentiated elements displayed moderate levels of CpG-methylation at the NRR ().
In order to compare the methylation levels of the CpGs in the NRR during germ cell development and in GCTs, average levels were calculated (). Here, the levels of CpG-methylation in the NRR of seminomas and embryonal carcinomas were comparable to fetal germ cells. This finding again strengthens the idea that GCTs arise from early fetal germ cells or fetal spermatogonia.Citation11,Citation23,Citation24 All other germ cell tumor entities display 40–80% of methylated CpGs at the NRR, most likely reflecting the various states of somatic differentiation of these tumors.
Next, we analyzed the methylation of CpGs in the NRR in various GCT-derived cell lines. The seminoma-like cell line TCam-2 and the embryonal carcinoma cell lines 2102EP, TERA1, TERA2 and NT2 displayed hypomethylation of the CpGs in the NRR, while two cell lines (833KE and NCCIT) displayed hypermethylation (). These results suggest that the methylation level of the CpGs in the NRR in GCT-cell lines remained unaltered in TCam-2, 2102EP, TERA1, TERA2 and NT2 during in vitro culture since they are identical to the methylation profile of the CpGs in the NRR of the GCT entities from which they originated. Of note, the CpGs in the NRR of 833KE, NCCIT and JKT-1 cells were found hypermethylated, suggesting that the methylation pattern had changed during adaptation of the cells to culture or during prolonged culture time. Alternatively, the data for JKT-1, together with experiments from us and others, further indicate that JKT-1 does not (or no longer) display seminoma-like characteristics.Citation25,Citation26 To address the question whether NANOG expression correlates to methylation of the CpGs in the NRR in GCTs and derived cell lines, we performed qRT-PCR analysis. NANOG expression levels were high in seminomas and embryonal carcinomas, while low and absent in teratomas, yolk sac tumors, choriocarcinomas and mixed non-seminomas (), indicating that methylation of the CpGs in the NRR correlates with NANOG expression in germ cell tumors.
Cell lines displaying a hypomethylated promoter region such as TERA1, TERA2, 2102EP, NT2, TCam-2 () showed NANOG expression. Accordingly, in GCT-derived cell lines with hypermethylated CpGs in the NRR (833KE, NCCIT and JKT-1) no or low level of NANOG expression was detected (). The qRT-PCR data shown were confirmed by RT-PCR and western blot analysis of NANOG expression in selected cell lines ( and D).
Discussion
In this study we convincingly show that a distinct promoter element (NRR; bp −306-bp -1) that harbors five CpG-dinucleotides is regulated by methylation and contributes to regulation of NANOG expression in human germ cells and germ cell tumors. Using a phylogenetic footprint, we demonstrate that location and number of CpG-dinucleotides are conserved only in humans and chimps, while the OCT3/4-SOX2 binding sites are detected in all species analyzed. This indicates, that the epigenetic modulation of OCT3/4-SOX2 driven NANOG expression by these five CpG-sequences might have evolved in the primate clade exclusively. In this context it is of interest that no SOX2 is expressed at any development stage of human germ cells, while this is found in those of murine species.Citation27 Here, we show that the NRR is able to drive luciferase expression in human cells positive for OCT3/4 and SOX2. The expression was markedly reduced after in vitro methylation of the NRR. However, in mice, other CpG islands within the Nanog promoter are subject to methylation resulting in epigenetic regulation of Nanog expression. There, the Nanog promoter is differentially methylated when murine ES cells are compared to trophoblast stem cellsCitation28 fibroblasts or cells from adult brain.Citation29 Using luciferase assays it was shown that the transcriptional activity of a proximal element correlates inversely to the level of its methylation.Citation28 Further, in mice it has been shown that the Oct3/4-Sox2 binding sites are functional as demonstrated by ChIP experiments. There, RNAi mediated knock-down of Oct3/4 and Sox2 expression led to a reduction in Nanog expression.Citation18 These data indicate that in humans DNA methylation of the five NRR CpGs is able to induce epigenetically silencing of NANOG expression and suggest that the region proximal to the transcriptional start site at the NANOG promoter is subject to epigenetic regulation in many species.
Next, we found that the NRR is hypomethylated in human fetal (c-KIT+) gonocytes where NANOG has been reported to be highly expressed.Citation5 Around week 24 of gestation transition of fetal gonocytes (c-KIT+) to fetal spermatogonia (MAGE-A4+) occurs.Citation21 MAGE-A4 positive cells display a hypomethylation of the NRR but lack NANOG expression. Since SOX2 is not expressed in MAGE-A4 positive germ cells of fetal human gonadsCitation13 and OCT3/4 expression is absent in c-KIT-, MAGE-A4+ fetal spermatogoniaCitation10 it is likely that NANOG downregulation in fetal spermatogonia is a consequence of a lack of transactivation via OCT3/4-SOX2 with no need of epigenetic silencing. Hypermethylation of the NRR can be observed in human sperm DNA and in cells from adult testicular tissue, suggesting that the epigenetic silencing marks at the NRR are established during maturation from spermatogonia to sperm. In summary, the NANOG expression correlates to NRR methylation in fetal germ cells, adult testis tissue and mature sperm. In spermatogonia NRR remains hypomethylated but NANOG is not expressed due to lack of OCT3/4-SOX2 expression. It is known that seminomas and c-KIT positive germ cells lack SOX2. Here, SOX17 might compensate for a lack of SOX2 expression transactivating NANOG in combination with OCT3/4.Citation13
Interestingly, global DNA methylation level analysis revealed hypermethylation of spermatogonia and hypomethylation of spermatids and sperm. A study on the overall methylation pattern of normal and malignant germ cells, detected using immunohistochemistry was recently published.Citation30 Using the same antibody, a hypomethylated pattern was found in male prenatal germ cells, which was gradually changed into a hypermethylated pattern during maturation into spermatogonia. This is in line with the observations presented here. Interestingly, indeed, the overall pattern of methylation, as found in the physiological germ cells is retained in the tumors derived from them. A global genome-wide promoter analysis of DNA methylation in mouse ES cells, embryonic germ (EG) cells, sperm, trophoblast stem cells and primary embryonic fibroblasts showed that the majority of the promoters found hypomethylated in ES and EG cells were also hypomethylated in sperm. In contrast, the NANOG promoter was highly methylated in sperm and hypomethylated in ES and EG cells.Citation31 These results in murine cells support the results reported here using human tissue. So, NRR-hypermethylation in sperm and in adult testes might be a way to epigenetically repress NANOG expression in order to control the pluripotency program and to prevent germ cell malignancies.
We demonstrate that GCTs and derived cell lines displaying a hypomethylated NRR express NANOG, while hypermethylated samples show no or very weak NANOG expression. Global demethylation resulted in an increase in NANOG expression, while NRR-specific methylation led to a significant repression of the NRR-activity.
Of note, two cell lines 833KE and NCCIT show a modest methylation of the NRR element but the level of NANOG expression is comparable to 2102EP cells, which display complete absence of methylation at the NRR. Either other promoter elements might override the repressive function of methylated CpGs within the NRR of 833KE and NCCIT cells or the level of OCT3/4 and SOX2 protein in 2102EP cells is low, allowing for moderate expression of NANOG only. This and other studies will help in further understanding the complex interrelation between epigenetic and genetic regulation of gene expression.
Materials and Methods
Sample handling and characterization.
The GCT samples were collected in the southwestern part of The Netherlands. Representative parts of the tumor were snap frozen in liquid nitrogen, or were fixed in 4% phosphate-buffered formalin for paraffin embedding. Diagnosis was carried out according to the World Health Organization classification.Citation14 We analyzed seminoma tissues (n = 6), embryonal carcinoma tissues (n = 5), yolk sac tumor tissues (n = 4), choriocarcinoma tissues (n = 2), teratoma tissues (n = 5) and mixed non-seminoma tissues (n = 4).
Cell culture.
TCam-2 cells were grown in RPMI supplemented with 10% fetal calf serum (FCS), 1% penicillin/streptomycin, 200 mM glutamine. 2102EP and NCCITCitation15 cells (obtained from Dr. F. Honecker, Hamburg University Medical Center, Department of Oncology/Haematology, Hamburg, Germany) were grown in Dulbecco's modified Eagle's medium:F12 medium (1:1) supplemented with 10% FCS, 1% penicillin/streptomycin, 200 mM glutamine. TERA-2 was grown in McCoy Medium plus 10% FCS, 1% penicillin/streptomycin, 200 mM glutamine. The cells were grown at 37°C and under 5% CO2. JKT-1 cells,Citation16 (obtained from Dr. Michiko Fukuda, The Burnham Institute, La Jolla, CA) were grown in minimal essential medium (MEM) plus 10% FCS, 1% penicillin/streptomycin, 200 mM glutamine.
Micro-dissection of fetal germ cells.
Testicular samples for micro-dissection were fixed in Bouin's solution and embedded in paraffin. Sections of 6 µm were mounted on membrane slides (P.A.L.M. Microlaser Technologies, Bernried, Germany) which were pretreated with poly-L-lysine and UV. All following steps were performed under sterile conditions and at room temperature. After de-paraffinization, slides were washed in washing buffer (0.1 M Tris, 0.1 M NaCl, pH 7.4) and pre-incubated in 5% bovine serum albumin (BSA) in washing buffer followed by immunohistochemical detection of MAGE-A4 1:100 (Spagnoli, Basel, Switzerland) or c-KIT 1:100 (A4502, DAKO, Hamburg, Germany) as described later. Slides were used for laser microdissection followed by laser pressure catapulting (LPC) with the MicroBeam system (P.A.L.M. Microlaser Technologies) to isolate c-KIT+/MAGE-A4+ fetal germ cells. We analyzed fetal germ cells (n = 3), fetal spermatogonia (n = 2) and spermatogonia of 5, 6, 9 and 33 year old patients (each n = 1).
DNA isolation.
DNA of freeze-dried tissue and micro-dissected from paraffin-embedded biopsies was isolated from germ cell tumors, normal testis tissue, fetal germ cells and germ cell tumor cell lines. DNA was isolated by adding 350 µl lysis-buffer (50 mM TRIs-HCl, pH 8; 100 mM EDTA; 100 mM NaCl; 1% SDS) and 40 µl Proteinase K (10 mg/ml) to the cell pellet. The cells were digested overnight at 55°C. The next day 150 µl NaCl (5 M) was added and cells were shaken for 5 min, followed by a centrifugation step (13,000 rpm) for 10 min. Supernatant was transferred to 500 µl isopropanol and centrifuged for 10 min at 13,000 rpm at 4°C. Precipitated DNA was washed twice with 500 µl 80% ethanol and air-dried. DNA was resolved in 200 µl water for 1 h at 55°C. Concentration and 260/280 nm, 260/230 nm purity ratios were determined by NanoDrop (Peqlab, Erlangen, Germany).
Protein isolation.
Proteins were isolated from TCam-2 cells, 2102EP cells and normal adult testis tissue by RIPA buffer 50 mM TRIS-HCl pH 8, 150 mM NaCl, 1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS + 1 tablet of protease inhibitor cocktail (Roche, Mannheim, Germany). The lysates were incubated for 30 min at 4°C, followed by a 20 min centrifugation step at 12,000 g, 4°C. The supernatant was used for western blot analysis. The protein concentration was determined by BCA Protein Assay Kit (Pierce, Rockford, IL).
5-Aza-2′-deoxycytidine treatment of JKT-1 cells.
JKT-1 cells were grown in MEM medium supplemented with 10% fetal calf serum (FCS), 1% penicillin/streptomycin, 200 mM glutamine and 200 µM 5-Aza-2′-deoxycytine (Sigma Aldrich, Taufkirchen, Germany) for 24 h, afterwards protein and RNA was isolated as described.
Western blot.
For protein analysis, we used the Mini-PROTEAN Electrophoresis Cell and Mini Trans-Blot system (BioRad, Munich, Germany). Protein was prepared by standard protocols and electrophoresed at 30 mA for 90 min. Gels were blotted onto a polyvinylidene fluoride membrane in a BioRad blotting chamber overnight at 30 V at 4°C according to published protocols. After the membrane had been blocked in PBSTM (phosphate-buffered saline, 0.1% v/v Tween 20, 5% low fat milk powder), it was incubated in primary antibodies raised against NANOG (N-17, sc-30331, 1:400, Santa Cruz) and b-actin (1:10,000) in PBSTM for 3 h at room temperature, followed by secondary antibodies (anti-rabbit-horseradish peroxidase [HRP], anti-mouse-HRP; all from DAKO, Hamburg, Germany) diluted 1:2,000. Finally, the membrane was incubated in 1 ml PierceSuper Signal West Pico chemiluminescent substrate (Perbio, Bonn, Germany) and the signal was detected by using Kodak X-Ray film (Kodak, Germany).
RT-PCR.
RNA of analyzed cell lines and tissues was extracted by TRIzol (Sigma, Germany) according to manufacturer's instructions. RNA quality was measured by NanoDrop (PeqLab, Erlangen, Germany). First-strand synthesis was performed according to Invitrogen's Super Script III manual (Invitrogen, Karlsruhe, Germany). 500 ng of total RNA were used for each first-strand synthesis. PCR was carried out at 95°C for 2 min, followed by 25 cycles of 94°C for 45 s, 58°C for 45 s and 72°C for 45 s. The following forward (F) and reverse (R) primers were used: NANOG RT F, 5′-GAT TTG TGG GCC TGA AGA AA-3′, NANOG RT R 5′-CAA AGG CAA ACA ACC CAC TT-3′, OCT3/4 RT F 5′-CGA AAG AGA AAG CGA ACC AG-3′, OCT3/4 RT R 5′-GCC GGT TAC AGA ACC ACA CT-3′, SOX2 RT F 5′-ATG CAC CGC TAC GAC GTG A-3′, SOX2 RT R 5′-CTT TTG CAC CCC TCC CAT T-3′.
Sodium bisulfite conversion and PCR.
To determine the methylation level of the different cell types, 500 ng DNA of each sample were treated with sodium bisulfite to convert all unmethylated cytosine-residues to uracil. The procedure was performed after the published protocol of the EpiTect Bisulfite Kit (Qiagen, Hilden, Germany). During PCR amplification the generated uracil-residues are converted to thymidine. Methylated cytosine-residues within a CpG-dinucleotide are left unaffected. 150 ng sodium-bisulfite treated DNA was used for each PCR reaction. PCR was carried out at 95°C for 2 min, followed by 40 cycles of 94°C for 45 s, 54°C for 45 s and 72°C for 45 s. The following primers, designed to recognize sodium bisulfite-treated DNA were used: NANOGP1BISF: 5′-TTA TAT TTT TGA TTT AAA AGT TGG AAA-3′, NANOGP1BISR: 5′-TAA CAT AAA ACA ACC AAC TCA ATC C-3′, OCT3/4PBISF 5′-ATT TGT TTT TTG GGT AGT TAA AGG T-3′, OCT3/4PBISR 5′-CCA ACT ATC TTC ATC TTA TAA CAT CC-3′.
Cloning and sequencing.
PCR products were ligated into pCR2.1 vector following the instructions of the TA-cloning Kit (Invitrogen, Karlsbad, Germany). The molecular ratio was 3:1 (insert:vector). A restriction digest was performed to check the isolated plasmids for the correct inserts. 1 µg Plasmid-DNA was digested with EcoR1 (1 U/µl) for 2 h. The reaction was stopped at 65°C for 20 min. The results were evaluated by agarose-gelelectrophoresis. The expected fragment for the NRR is about 375 bp in size (360 bp NANOG promoter region +∼15 bp pCR2.1 backbone fragments). Plasmids were sequenced by Entelechon GmbH (Regensburg, Germany). For each tumor entity 2–7 different samples were analyzed. Each sample was sequenced sixfold. 1 µg genomic TCam-2 DNA was treated for 4 h with M.SssI methylase (4 U/µl, NEB) as positive control following manufacturers protocol. The NRR fragment used in the luciferase assays was amplified by PCR (primers included KpnI and NheI recognition sites), ligated into pGL3basic vector (Promega, Mannheim, Germany) after KpnI/NheI (NEB, Frankfurt, Germany) restriction and purification by T4 Ligase (NEB, Frankfurt, Germany).
Quantitative RT-PCR.
High-quality total RNA was extracted from patient-derived germ cell tumor samples and cell-lines using an RNAqueous-4PCR kit (Ambion/Applied Biosystems, Nieuwerkerk a/d IJssel, the Netherlands) according to the manufacturer's instructions. Samples were treated with DNase and checked for residual DNA contamination by PCR. Quantitative PCR was performed using the 7900HT Fast Real-Time PCR System (Applied Biosystems). For the detection of NANOG expression, the following primer-set was used: NANOG-F: 5′-CGG AGA CTG TCT CTC CTC TTC C-3′ and NANOG-R: 5′-GGT AGG TGC TGA GGC CTT CTG C-3′. Quantitative values were obtained from the Ct. NANOG mRNA was quantified relative to HPRT [NANOG mRNA value = 2(mean Ct HPRT - Ct NANOG )].
Immunohistochemical staining of fetal germ cells.
Testes were dissected, fixed in 4% phosphate-buffered formalin for 2 days at room temperature and processed in paraffin. Dewaxed, 4 µm thick tissue sections were microwave pretreated (3 × 5 min, 600W) in Tris-EDTA buffer (10 mmol/l tris base, 1 mmol/l EDTA solution, 0.05% Tween 20, pH 8.0). Primary antibodies were used in the dilutions given to detect the following proteins: MAGE-A4 1:100 (generous gift of Dr. G. Spagnoli, University of Basel, Basel, Switzerland), c-KIT 1:100 (A4502, DAKO, Hamburg, Germany), OCT3/4 1:200 (c-10, Santa Cruz, Heidelberg, Germany), 5-Methyl-Cytidine 1:200 (ab10805, Abcam, Cambridge, UK) and SOX2 1:200 (245610, R&D Systems, Wiesbaden, Germany). Immunohistochemistry was performed using the Dako EnVision-AEC Kit according to manufacturer's protocol (Dako, Hamburg, Germany). Briefly, endogenous peroxidase was blocked for 5 min in 0.03% hydrogen peroxide (diluted in distilled water). Sections washed in Tris-buffered saline (0.05 mol/l Tris and 0.85% NaCl, pH 7.6) were incubated with primary antibodies overnight at 4°C. Thereafter, a horseradish peroxidase (HRP)-labeled polymer conjugated with a secondary antibody was applied (Dako EnVision-AEC Kit). The staining was visualized with 3-amino-9-ethyl-carbazole and counterstained with hematoxylin. Negative controls were performed using buffer instead of the primary antibody. Additional negative controls were performed by incubation with pre-immune serum from each animal species (rabbit, mouse, goat) instead of a primary antibody. All controls resulted in complete absence of an immunohistochemical signal.
Luciferase assay.
pGL3basic vector (Promega, Mannheim, Germany) containing the luciferase gene under control of the NRR was transfected in 4 · 104, 2102EP cells at a ratio of 5:2 (µl transfection reagent: µg DNA). FuGeneHD (Roche, Germany) was used as transfection reagent. Additionally a pGL3basic-NRR construct methylated by M.SssI and empty pGL3 vector was transfected. Furthermore, a pGL3basic-NRR with mutated OCT3/4-SOX2 binding sites (original OCT3/4-SOX2 binding sites: CTT TTG CAT TAC AAT G; mutated OCT3/4-SOX2 binding sites: CTT TCA AAT TAA CCT G) and a pGL3basic-NRR construct with mutated OCT3/4-SOX2 bindings sites methylated by M.SssI was transfected. Untreated 2102EP cells were used as additional controls. 48 h later, cells were lysed for 5 min in 100 µl 1x lysis buffer (5x: 125 mM Tris pH 7.8, 10 mM EDTA, 100 mM DTT, 50% glycerol, 5% TritonX-100). After centrifugation (5′, 14,000 rpm, 4°C) 10 µl of the supernatant were transferred to a 96-well plate. Luciferase activity was measured one second after addition of the luciferase-assay buffer (20 mM tricine, 1.07 mM (MgCO3), 4Mg(OH)2•5H2O, 2.67 mM MgSO4, 0.1 mM EDTA, 33.3 mM DTT, 270 µM Coenzyme A, 530 µM ATP, 470 µM Luciferin) by a luminometer. Values were normalized against individual protein levels, measured by BCA assay (BCA Protein Assay KIT, Pierce, Rockford, IL).
Figures and Tables
Figure 1 Luciferase assay in 2102EP embryonal carcinoma cells. (A) Relative luciferase activity of the NRR (pGL3-NRR), the in vitro methylated NRR (pGL3-NRR M.SssI), the NRR with mutated OCT3/4-SOX2 binding site (mOS) and in vitro methylated NRR with mutated OCT3/4-SOX2 binding site (mOS M.SssI). Untransfected (2102EP u) and “empty” vector transfected cells (pGL3basic) served as controls. (B) NRR methylation level in JKT-1 cells before (untreated) and after treatment with 10 µM 5-aza-2'-deoxycytidine (5-aza). Filled circles indicate methylated CpGs, empty circles indicate demethylated CpGs. (C) RT-PCR analysis of NANOG-expression before (untreated) and after treatment with 5-aza (5-aza).
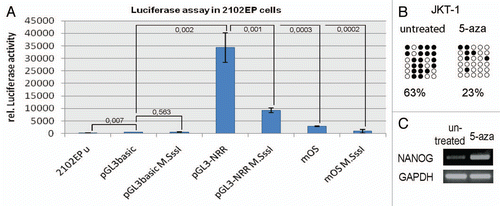
Figure 2 (A and B) Immunohistochemical staining of fetal germ cells for c-KIT (A) and MAGE-A4 (B). Arrowheads indicate cells positive for the respective protein. Stained cells were extracted by laser micro-dissection for DNA isolation and sodium-bisulfite sequencing. Scale bar: 500 µm. (C–G) NRR DNA-methylation pattern of germ cells at developmental stages indicated. Adult spermatogonia (E) were isolated by laser micro-dissection. DNA of adult testis tissues (F) was isolated from whole tissue lysates, while sperm DNA was isolated from ejaculates (G). (H) Positive control (M.SssI treated DNA from TCam-2 cells). Filled circles indicate methylated CpGs, empty circles indicate demethylated CpGs.
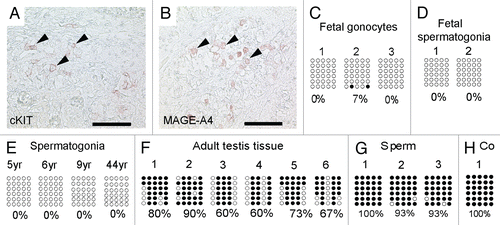
Figure 3 Immunohistochemical staining of testis sections at week 19 (A and C) and 24 (B and D) of gestation for OCT3/4 (A and B) and SOX2 (C and D). Arrows indicate cells positive for OCT3/4. Scale bar: 50 µm.
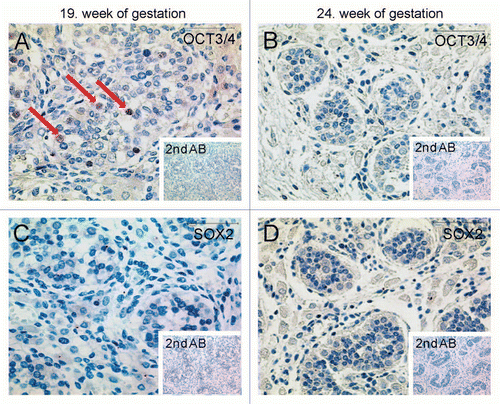
Figure 4 Immunohistochemical staining detecting global DNA-methylation in different testis samples using 5-mC antibody. (A) Red arrows point at spermatogonia of a testis from 2-year-old patient being positive for 5 mC antibody. (B) Testis section of a 6-year-old patient shows global hypermethylation of spermatogonia, too. (C and D) Spermatids and sperm of donors age 22 and 29 show global hypomethylation (arrowheads). Scale-bars: A and B: 200 µm; C and D: 50 µm.
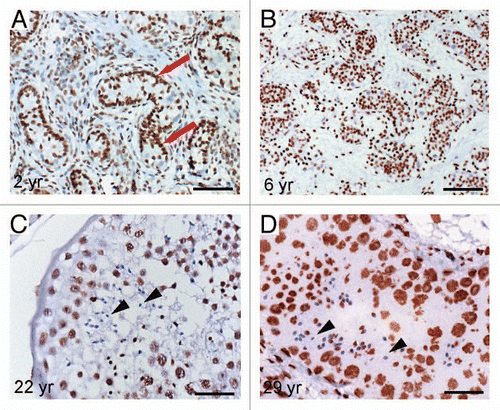
Figure 5 Graphical illustration of the NRR DNA-methylation pattern in indicated GCT samples (A–F). Filled circles indicate methylated CpGs, empty circles indicate demethylated CpGs. (G) Diagram of relative NRR methylation levels of germ cells and GCTs.
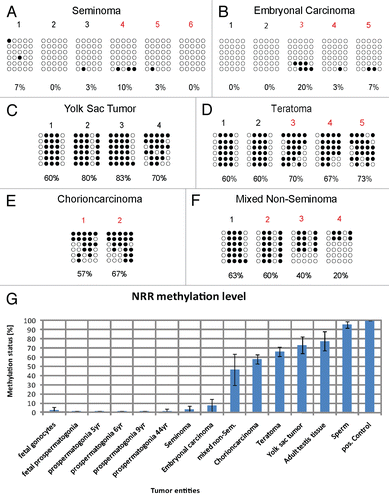
Figure 6 (A) NRR-methylation analysis of GCT cell lines. Filled circles indicate methylated CpGs, empty circles indicate demethylated CpGs. (B) qRT-PCR anaylsis of NANOG expression in GCT and control tissues (red colored samples in ) and corresponding cell lines. (C) RT-PCR and (D) western blot of cell lines indicated detecting expression and protein level of NANOG.
Additional material
Download Zip (2.2 MB)Acknowledgements
This work was supported by a grant from the DFG to H.S. (Scho 503/9). We thank Dr. Janet Shipley (Institute of Cancer Research, Sutton, England) for the TCam-2 cells, Dr. Michiko Fukuda (The Burnham Institute, La Jolla, CA.) for the JKT-1 cells and Dr. F. Honecker (Hamburg University Medical Center, Department of Oncology/Hematology, Hamburg, Germany) for the EC cell line 2102EP.
References
- Ghoshal K, Datta J, Majumder S, Bai S, Kutay H, Motiwala T, et al. 5-Aza-deoxycytidine induces selective degradation of DNA methyltransferase 1 by a proteasomal pathway that requires the KEN box, bromo-adjacent homology domain and nuclear localization signal. Mol Cell Biol 2005; 25:4727 - 4741
- Muhlhauser J, Crescimanno C, Kasper M, Zaccheo D, Castellucci M. Differentiation of human trophoblast populations involves alterations in cytokeratin patterns. J Histochem Cytochem 1995; 43:579 - 589
- Chambers I, Colby D, Robertson M, Nichols J, Lee S, Tweedie S, et al. Functional expression cloning of Nanog, a pluripotency sustaining factor in embryonic stem cells. Cell 2003; 113:643 - 655
- Mitsui K, Tokuzawa Y, Itoh H, Segawa K, Murakami M, Takahashi K, et al. The homeoprotein Nanog is required for maintenance of pluripotency in mouse epiblast and ES cells. Cell 2003; 113:631 - 642
- Hoei-Hansen CE, Almstrup K, Nielsen JE, Brask Sonne S, Graem N, et al. Stem cell pluripotency factor NANOG is expressed in human fetal gonocytes, testicular carcinoma in situ and germ cell tumours. Histopathology 2005; 47:48 - 56
- Hart AH, Hartley L, Parker K, Ibrahim M, Looijenga LH, Pauchnik M, et al. The pluripotency homeobox gene NANOG is expressed in human germ cell tumors. Cancer 2005; 104:2092 - 2098
- Cavaleri F, Scholer HR. Nanog: a new recruit to the embryonic stem cell orchestra. Cell 2003; 113:551 - 552
- Hyslop L, Stojkovic M, Armstrong L, Walter T, Stojkovic P, Przyborski S, et al. Downregulation of NANOG induces differentiation of human embryonic stem cells to extraembryonic lineages. Stem Cells 2005; 23:1035 - 1043
- de Sousa Lopes SM, Hayashi K, Surani MA. Proximal visceral endoderm and extraembryonic ectoderm regulate the formation of primordial germ cell precursors. BMC Dev Biol 2007; 7:140
- Gaskell TL, Esnal A, Robinson LL, Anderson RA, Saunders PT. Immunohistochemical profiling of germ cells within the human fetal testis: identification of three subpopulations. Biol Reprod 2004; 71:2012 - 2021
- Oosterhuis JW, Looijenga LH. Testicular germ-cell tumours in a broader perspective. Nat Rev Cancer 2005; 5:210 - 222
- Biermann K, Zhou H, Buttner R. [Molecular pathology of testicular germ cell tumors: an update]. Pathologe 2008; 29:348 - 353
- de Jong J, Stoop H, Gillis AJ, van Gurp RJ, van de Geijn GJ, Boer M, et al. Differential expression of SOX17 and SOX2 in germ cells and stem cells has biological and clinical implications. J Pathol 2008; 215:21 - 30
- Mostofi FK, Sesterhenn IA. Pathology of germ cell tumors of testes. Prog Clin Biol Res 1985; 203:1 - 34
- Josephson R, Ording CJ, Liu Y, Shin S, Lakshmipathy U, Toumadje A, et al. Qualification of embryonal carcinoma 2102Ep as a reference for human embryonic stem cell research. Stem Cells 2007; 25:437 - 446
- Kinugawa K, Hyodo F, Matsuki T, Jo Y, Furukawa Y, Ueki A, et al. Establishment and characterization of a new human testicular seminoma cell line, JKT-1. Int J Urol 1998; 5:282 - 287
- Kuroda T, Tada M, Kubota H, Kimura H, Hatano SY, Suemori H, et al. Octamer and Sox elements are required for transcriptional cis regulation of Nanog gene expression. Mol Cell Biol 2005; 25:2475 - 2485
- Rodda DJ, Chew JL, Lim LH, Loh YH, Wang B, Ng HH, et al. Transcriptional regulation of nanog by OCT4 and SOX2. J Biol Chem 2005; 280:24731 - 24737
- Freberg CT, Dahl JA, Timoskainen S, Collas P. Epigenetic reprogramming of OCT4 and NANOG regulatory regions by embryonal carcinoma cell extract. Mol Biol Cell 2007; 18:1543 - 1553
- Deb-Rinker P, Ly D, Jezierski A, Sikorska M, Walker PR. Sequential DNA methylation of the Nanog and Oct-4 upstream regions in human NT2 cells during neuronal differentiation. J Biol Chem 2005; 280:6257 - 6260
- Pauls K, Schorle H, Jeske W, Brehm R, Steger K, Wernert N, et al. Spatial expression of germ cell markers during maturation of human fetal male gonads: an immunohistochemical study. Hum Reprod 2006; 21:397 - 404
- Almstrup K, Hoei-Hansen CE, Wirkner U, Blake J, Schwager C, Ansorge W, et al. Embryonic stem cell-like features of testicular carcinoma in situ revealed by genome-wide gene expression profiling. Cancer Res 2004; 64:4736 - 4743
- Sonne SB, Almstrup K, Dalgaard M, Juncker AS, Edsgard D, Ruban L, et al. Analysis of gene expression profiles of microdissected cell populations indicates that testicular carcinoma in situ is an arrested gonocyte. Cancer Res 2009; 69:5241 - 5250
- Bussey KJ, Lawce HJ, Himoe E, Shu XO, Heerema NA, Perlman EJ, et al. SNRPN methylation patterns in germ cell tumors as a reflection of primordial germ cell development. Genes Chromosome Canc 2001; 32:342 - 352
- de Jong J, Stoop H, Gillis AJ, Hersmus R, van Gurp RJ, van de Geijn GJ, et al. Further characterization of the first seminoma cell line TCam-2. Genes Chromosome Canc 2008; 47:185 - 196
- Eckert D, Nettersheim D, Heukamp LC, Kitazawa S, Biermann K, Schorle H. TCam-2 but not JKT-1 cells resemble seminoma in cell culture. Cell Tissue Res 2008; 331:529 - 538
- Avilion AA, Nicolis SK, Pevny LH, Perez L, Vivian N, Lovell-Badge R. Multipotent cell lineages in early mouse development depend on SOX2 function. Genes Dev 2003; 17:126 - 140
- Hattori N, Imao Y, Nishino K, Hattori N, Ohgane J, Yagi S, et al. Epigenetic regulation of Nanog gene in embryonic stem and trophoblast stem cells. Genes Cells 2007; 12:387 - 396
- Blelloch R, Wang Z, Meissner A, Pollard S, Smith A, Jaenisch R. Reprogramming efficiency following somatic cell nuclear transfer is influenced by the differentiation and methylation state of the donor nucleus. Stem Cells 2006; 24:2007 - 2013
- Wermann H, Stoop H, Gillis AJ, Honecker F, van Gurp RJ, Ammerpohl O, et al. Global DNA methylation in fetal human germ cells and germ cell tumours: association with differentiation and cisplatin resistance. J Pathol 2010; 221:433 - 442
- Farthing CR, Ficz G, Ng RK, Chan CF, Andrews S, Dean W, et al. Global mapping of DNA methylation in mouse promoters reveals epigenetic reprogramming of pluripotency genes. PLoS Genet 2008; 4:1000116