Abstract
Alterations in the epigenetic programming of sex differences in the brain may underlie sexually dimorphic neurodevelopmental disorders. Sex differences have been found in DNA methyltransferases 3a, DNA methylation patterns, MeCP2, and nuclear corepressor within the developing brain. Natural variations in these epigenetic mechanisms have profound consequences on gene expression and brain function. Exogenous or endogenous perturbations during development may impact these epigenetic processes and alter the trajectory of the developing brain and confer sexually dimorphic risk and resilience for developing a neurological or mental health disorder.
Sexual differentiation of the neonatal rat brain is regulated by dynamic processes occurring at the level of DNA, resulting in sexually dimorphic gene expression. Steroid hormone receptors act partly in the developing brain by recruiting co-regulatory proteins and other transcription factors that lead to altered gene expression. Recent data indicate that epigenetic processes play an important role in sexual differentiation of the brain. Methylation of DNA is an epigenetic process that can alter gene expression without altering the original DNA sequence. Methylation marks on DNA can recruit methyl-binding proteins (MBPs), which in turn may recruit co-repressor proteins and histone deacetylase complexes (HDACs) to modify chromatin and ultimately alter gene transcription. The dynamic interaction between epigenetic factors and chromatin leads to functional plasticity within the brain that may explain the diversity of responses to endogenous and exogenous signals. Subtle variations in these epigenetic factors may also partially explain individual differences in the development of neurological and mental health disorders. We propose that sex differences in these epigenetic factors not only contribute to sexual differentiation of the brain and social behavior, but that they may confer sexually dimorphic risk and resilience for developing neurological and mental health disorders later in life ().
Sex Differences in Epigenetic Mechanisms
DNA methyltransferases and brain sex differences.
Methylation of DNA occurs when a methyl group attaches to a cytosine within a 5′-CpN-3′ dinucleotide site through an enzymatic reaction that is catalyzed by DNA cytosine-5-methyltransferases (DNMTs). While this bond can be rather stable, it has also been shown to be reversible.Citation1 DNA methylation occurs mostly at CpG sites, with lower levels at CpA, CpC and CpT sites.Citation2,Citation3 DNMTs include DNMT1, DNMT2, DNMT3a, DNMT3b, and DNMT3L, which appear to differ in function. DNMT1 is considered important in maintaining DNA methylation patterns due to its high preference for hemimethylated DNA,Citation4,Citation5 whereas DNMT3a and DNMT3b are important in de novo methylation.Citation6,Citation7 Some data suggest that DNMT3a protein expression increases during the first three weeks of postnatal brain development and then declines, whereas DNMT3b protein expression is undetectable during this time period,Citation8 suggesting that DNMT3a may be more important in inducing de novo methylation in postnatal brain.
Interestingly, there appears to be a sex difference in the levels of DNMT3a within the developing amygdala.Citation9 Specifically, females express significantly more DNMT3a at postnatal day 1 (PN1) in the amygdala, but not within the preoptic area or medial basal hypothalamus. This sex difference is eliminated by the second week of life, suggesting a small window in which some genes may be differentially marked between males versus females. Sex differences in hormone exposure during early postnatal development appear partly responsible for the sex difference in DNMT3a levels, as treatment with either estradiol benzoate (EB) or dihydrotestosterone (DHT) decreases DNMT3a levels within the amygdala. This is particularly relevant as differences in hormones levels due to endogenous or exogenous sources, such as early-life adversity, may program lasting differences in brain function by modifying the expression levels of DNMTs within the developing amygdala.
As the amygdala is known to be important for modulating juvenile social play,Citation10 aggression,Citation11 and anxiety,Citation12,Citation13 programming or possibly reprogramming gene expression within the amygdala is likely to impact numerous socio-emotional functions. While the role of DNMT3a on programming sex differences in the amygdala has not been directly tested, recent studies support a role for DNMTs in regulating emotional behavior and synapse function, as well as DNA methylation patterns.Citation14,Citation15 Changes seen in DNMT3a, but not DNMT1, expression during the early postnatal period suggest that DNMT3a may be a key factor in producing sex differences in methylation patterns during brain development and may induce methylation patterns that alter the probability of a neurodevelopment disorder.
DNA methylation: ERα promoter methylation and brain sex differences.
As DNMTs levels are sexually dimorphic and responsive to changes in endogenous steroid hormone levels, it is not surprising that recent data indicate that DNA methylation itself is sexually dimorphic. Previous data have shown that variations in maternal care can lead to altered DNA methylation patterns.Citation16,Citation17 In particular, differences in maternal care alter ERα promoter methylation patterns in the developing offspring's brain and these differences correlate with changes in ERα expression.Citation16 Interestingly, mother rats differentially groom male versus female offspring,Citation18,Citation19 and thereby may be modifying the DNA methylation patterns in a sex-specific manner. Indeed, it was recently found that there is a sex difference in DNA methylation of the ERα promoter region within the developing rat preoptic areaCitation20 and amygdala.Citation21 Specifically, we found that males exhibit higher levels of methylation within the 5′ flanking region of ERα exon 1b promoter region contrasted to females during the first week of postnatal life.Citation20 These data are consistent with previous findings that males express lower levels of ERα mRNA compared to females within the developing brain.Citation22–Citation24 Perhaps more importantly, treatment of newborn female rats with estradiol reprogrammed ERα promoter methylation patterns, suggesting that differences in hormone exposure within the first few days of life can have lasting consequences on gene expression levels by altering DNA methylation marks within the developing brain.Citation20
To further clarify the contributions of maternal care to sex difference in ERα promoter methylation and expression, we modeled a component of maternal care using a paradigm called simulated maternal grooming (SMG) to mimic the somatosensory stimuli associated with maternal grooming. This paradigm controls for other factors of maternal care and focuses on the tactile stimulation of licking and grooming. We found that this subtle somatosensory stimulus altered DNA methylation patterns within the developing brain, in particular within the developing preoptic areaCitation20 and amygdala.Citation21 That is, we find sex differences in DNA methylation patterns within the developing brain, and these differences, in part, appear to result from differences in maternal care directed at male vs. female offspring. We find that providing females with additional somatosensory stimuli associated with maternal grooming produces male-typical patterns of ERα mRNA levels and methylation of certain CpG sites in the exon 1b ERα promoter region. As the amygdala is a critical site for socio-emotional behavior, these data suggest that differences in mother-infant interactions may modify social or emotional processes by reprogramming DNA methylation patterns within the developing amygdala.
Intriguingly, the modification of ERα promoter methylation patterns by maternal stimuli is not immediate and appears to be established gradually over the first ten days of neonatal life. That is, we found that SMG-induced ERα methylation is established between P2 and P10. It is not clear if there is a critical threshold in the amount of environmental stimulus received to adjust DNA methylation patterns or if it may be time dependent; however, these data do suggest that the programming of ERα methylation is tightly regulated during development. Another possibility is that the connections relaying somatosensory stimuli from the periphery to the brain may not be fully established during the first few days of life and, therefore, those connections cannot trigger processes controlling DNA methylation. Any of these possibilities suggest a critical window during early neonatal development in which subtle variations in maternal care could reprogram DNA methylation patterns; however, it is also plausible that an accumulation of methylation marks in a number of cells are needed to detect a change in methylation pattern within that brain region. We suggest a critical window as the functional meaning of somatosensory stimuli is likely to decrease with age and therefore, are no longer able to reprogram ERα promoter methylation. However, this is not to suggest that other factors are constrained to this same time period or cannot override this window. For example, it is likely that the consequences of maternal neglect are less confined to this potential developmental window. It is also likely that other signals can later reverse these patterns later in life. A recent report indicates that some of the same CpG sites programmed during development can be altered by circulating steroid hormone levels in the adult brain.Citation25 This suggests some DNA markings are not as stable as previously thought, and that some of these patterns need to be maintained by an endogenous or exogenous signal later in life. This observed reversibility of DNA methylation questions the stability of methylation patterns within particular regions of DNA, at least on single CpG sites located within a promoter region.
Methyl-CpG binding proteins: MeCP2 brain sex differences and function.
It is likely that binding of methyl-CpG binding proteins to methylated DNA is essential in reading methylation marks that alter gene expression. A family of methyl-CpG-binding proteins was first discovered after the characterization of the methyl-CpG-binding domain (MBD) that binds to the methylated DNA.Citation26 Members of methyl-CpG-binding proteins include Kaiso, MBD1, MBD2, MBD3, MBD4 and Methyl-CpG-binding protein 2 (MeCP2). Upon binding to methylated DNA, methyl-binding proteins (MBPs) recruit co-repressor proteins and HDACs to modify chromatin and repress gene transcription.
MeCP2 was the first methyl-CpG binding protein to be described that binds methylated CpG sites.Citation27 Importantly, mutations in MECP2, an X-linked gene, are believed to cause Rett syndrome, a progressive neurodevelopmental disorder.Citation28 Rett syndrome is diagnosed mainly in females, as it is generally lethal in males. Mutations of MECP2 in mice appear to recapitulate many of the neurological symptoms occurring in Rett syndrome,Citation29,Citation30 and a recent study reports that re-expression of MeCP2 in these mice results in a reversal of many of these symptoms.Citation31 These data indicate the importance of MeCP2 in the etiology of Rett syndrome. More recent reports have suggested that subtle reductions in MeCP2 expression along with aberrant MECP2 methylation patterns occur in individuals with autism,Citation32 another sexually dimorphic neurodevelopmental disorder. In contrast to Rett syndrome, autism is known to occur at higher rates in boys. Together, this could suggest that the male brain is less able to compensate for reductions or loss of MeCP2.
In the rodent brain, females express higher levels of MeCP2 mRNA within the developing amygdala compared to males.Citation33 It is possible that there may be a critical threshold of MeCP2 expression, and that reduction under this threshold may be detrimental to brain development. Indeed, transiently reducing MeCP2 expression within the amygdala disrupted the organization of juvenile social play behavior in males, but not females.Citation34 It is important to note that disruption of MeCP2 in males did not alter their performance in a sociability task, suggesting that these males were less apt to engage in active social interactions that are typical of juvenile male rodents. These data not only support the concept that epigenetic factors, such as methyl-binding proteins, contribute to sexual differentiation of the brain and social behavior, but that disruption in its expression may induce sexually dimorphic behavioral disruptions later in life. These data could suggest a potential role for MeCP2 in the etiology of autism spectrum disorders, and do support the idea that the male brain is less able to adapt to reductions in MeCP2 levels.
Co-repressor complexes: NCor sex differences and function.
Methyl-CpG binding proteins bind to methylated DNA and can initiate the recruitment of nuclear co-repressors and histone deacetylase to form repressor complexes. These repressor complexes can inhibit gene expression by removing acetyl groups from histones leading to condensation of the chromatin and gene repression.Citation35 Co-repressor complexes recruited by methyl-binding proteins include multi-protein Sin3, NuRD, CoREST, and the NCoR/SMRT repressor complexes.Citation36 It is likely that different combinations of these co-repressor complexes allows for increased diversification of function and thereby phenotypical variations.
NCoR was one of the first corepressors to be identified through its interaction with thyroid hormone receptors;Citation37 however, it was also reported to interact with androgen receptors,Citation38,Citation39 estrogen receptorsCitation40 and progestin receptors.Citation41 NCoR has also been shown recently to interact directly, as well as indirectly, with methyl-binding proteins.Citation42–Citation45 In particular, NCoR is reported to interact with Kaiso, MeCP2 and other methyl-binding proteinsCitation42–Citation44 and may interact indirectly with methyl-binding proteins through its association with the Sin3 co-repressor complex.Citation45
While the functional role of co-repressors in the developing brain is still being elucidated, recent data indicate that there is a sex difference in NCoR levels within the developing rat amygdala. Specifically, females express higher levels of NCoR mRNA than do males. These differences may be partly due to differences in hormone exposure, as treatment with EB decreased the expression of NCoR in females to male like levels.Citation46 This is interesting as differences in hormones levels due to endogenous or exogenous sources, such as early-life adversity, may alter NCoR expression levels within the developing amygdala and thereby change the cell or tissue's response to further cues. A transient, targeted disruption of NCoR within the developing amygdala was found to have lasting consequences on both juvenile social play and anxiety-like behaviors. While the alterations in juvenile social play occurred only in males, the transient reduction of NCoR had lasting consequences on increasing anxiety-like behavior in both juvenile males and females. These data indicate that transient disruptions in corepressors complexes within the developing brain can have sexually dimorphic consequences on juvenile social behavior and have alterations in anxiety-like behavior in both sexes. It is important to consider that sex differences in epigenetic processes may not always induce sex differences, but may be there to reduce sex differences. This is best illustrated with the NCoR data in which lowering NCoR expression levels within the developing amygdala further enhanced male-typical juvenile play behavior in males, but to atypical levels.Citation46 Therefore, NCoR may play a role in reducing sex differences in areas of the brain where sex differences could be detrimental.
Conclusions
Our laboratory has found that the amygdala exhibits sex differences in multiple epigenetic factors during brain development, and these factors impact later behavior. Specifically, we found that neonatal females express significantly higher levels of MeCP2, NCoR and DNMT3a than males within the amygdala. We have also found that transiently decreasing the expression of genes that are recruited to methylated DNA, such as MeCP2 and NCoR, within the amygdala during the neonatal period results in altered social behavior later in life.Citation34,Citation46 Interestingly, transient reductions in MeCP2 within the developing amygdala decreased juvenile social play in males to female levels, while transient reductions in NCoR hyper-masculinized juvenile social play in males. These data suggest that different epigenetic factors or their combinations modify divergent phenotypes. It is intriguing that males express significantly less DNMT3a, MeCP2 and NCoR within the developing amygdala. While it is not clear why this pattern of lowered repression exist, it could suggest that the male amygdala is undergoing active differentiation induced by steroid hormone exposure during that time period. This potential increased window of differentiation of the male amygdala could put males at risk if a perturbation occurs during this time of brain organization.
Understanding epigenetic mechanisms, which underlie sexually dimorphic gene expression and function, is critical for further understanding both typical and atypical neurodevelopment. Furthermore, sexual differentiation of the brain is a powerful model to study epigenetic mechanisms, as brief exposure to steroid hormones can reorganize the brain and have lasting consequences on behavior. Perhaps more important, studying the male versus female brain provides a natural model of risk or resilience for some mental health disorders. For example, autism spectrum disorders, Rett syndrome, attention deficit hyperactivity disorder and schizophrenia, have sex differences in prevalence, time of onset and/or severity. As these disorders are believed to have an epigenetic component, sex differences in epigenetic processes may alter ones risk or resilience to develop a particular disorder. Therefore, it will be very interesting to further examine sex differences in epigenetic mechanisms and how these differences contribute to typical differentiation of the brain and social behavior, but also how they may confer sexually dimorphic risk or resilience for developing neurological and mental health disorders.
Figures and Tables
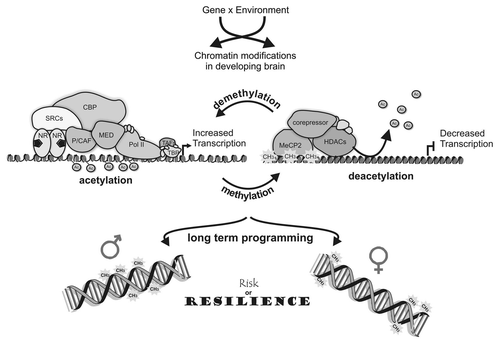
Figure 1 Sex differences in epigenetic mechanisms underlie risk and resilience for mental health disorders. Gene and environmental interactions remodel chromatin structure by recruiting co-regulatory proteins and other transcription factors resulting in either increased or decreased gene transcription rates. These interactions can lead to methylation or demethylation of DNA, as well as acetylation or deacetylation of histones. Lasting chromosomal alterations that impact gene and brain function can lead to gender specific altered risk or resilience to neurodevelopmental disorders later in life.
References
- Métivier R, Gallais R, Tiffoche C, Le Péron C, Jurkowska RZ, Carmouche RP, et al. Cyclical DNA methylation of a transcriptionally active promoter. Nature 2008; 452:45 - 50
- Grafstrom RH, Yuan R, Hamilton DL. The characteristics of DNA methylation in an in vitro DNA synthesizing system from mouse fibroblasts. Nucleic Acids Res 1985; 13:2827 - 2842
- Ramsahoye BH, Biniszkiewicz D, Lyko F, Clark V, Bird AP, Jaenisch R. Non-CpG methylation is prevalent in embryonic stem cells and may be mediated by DNA methyltransferase 3a. Proc Natl Acad Sci USA 2000; 97:5237 - 5242
- Pradhan S, Bacolla A, Wells RD, Roberts RJ. Recombinant human DNA (cytosine-5) methyltransferase. I. Expression, purification and comparison of de novo and maintenance methylation. J Biol Chem 1999; 274:33002 - 33010
- Yoder JA, Soman NS, Verdine GL, Bestor TH. DNA (cytosine-5)-methyltransferases in mouse cells and tissues. Studies with a mechanism-based probe. J Mol Biol 1997; 270:385 - 395
- Okano M, Xie S, Li E. Cloning and characterization of a family of novel mammalian DNA (cytosine-5) methyltransferases. Nat Genet 1998; 19:219 - 220
- Hsieh CL. In vivo activity of murine de novo methyltransferases, Dnmt3a and Dnmt3b. Mol Cell Biol 1999; 19:8211 - 8218
- Feng J, Chang H, Li E, Fan G. Dynamic expression of de novo DNA methyltransferases Dnmt3a and Dnmt3b in the central nervous system. J Neurosci Res 2005; 79:734 - 746
- Kolodkin MH, Auger AP. Sex difference in the expression of DNA methyltransferase 3a (DNMT3a) in the rat amygdala during development. J Neuroendocrinol 2011; In press
- Meaney MJ, Dodge AM, Beatty WW. Sex-dependent effects of amygdaloid lesions on the social play of prepubertal rats. Physiol Behav 1981; 26:467 - 472
- Pinel JP, Treit D, Rovner LI. Temporal lobe aggression in rats. Science 1977; 197:1088 - 1089
- Hitchcock J, Davis M. Lesions of the amygdala, but not of the cerebellum or red nucleus, block conditioned fear as measured with the potentiated startle paradigm. Behav Neurosci 1986; 100:11 - 22
- Shibata K, Kataoka Y, Yamashita K, Ueki S. An important role of the central amygdaloid nucleus and mammillary body in the mediation of conflict behavior in rats. Brain Res 1986; 372:159 - 162
- LaPlant Q, Vialou V, Covington HE 3rd, Dumitriu D, Feng J, Warren BL, et al. Dnmt3a regulates emotional behavior and spine plasticity in the nucleus accumbens. Nat Neurosci 2010; 13:1137 - 1143
- Feng J, Zhou Y, Campbell SL, Le T, Li E, Sweatt JD, et al. Dnmt1 and Dnmt3a maintain DNA methylation and regulate synaptic function in adult forebrain neurons. Nat Neurosci 2010; 13:423 - 430
- Champagne FA, Weaver IC, Diorio J, Dymov S, Szyf M, Meaney MJ. Maternal care associated with methylation of the estrogen receptor-alpha1b promoter and estrogen receptor-alpha expression in the medial preoptic area of female offspring. Endocrinology 2006; 147:2909 - 2915
- Weaver IC, Cervoni N, Champagne FA, D'Alessio AC, Sharma S, Seckl JR, et al. Epigenetic programming by maternal behavior. Nat Neurosci 2004; 7:847 - 854
- Moore CL, Morelli GA. Mother rats interact differently with male and female offspring. J Comp Physiol Psychol 1979; 93:677 - 684
- Moore CL. Maternal contributions to the development of masculine sexual behavior in laboratory rats. Dev Psychobiol 1984; 17:347 - 356
- Kurian JR, Olesen KM, Auger AP. Sex differences in epigenetic regulation of the estrogen receptor-alpha promoter within the developing preoptic area. Endocrinology 2010; 151:2297 - 2305
- Edelmann MN, Auger AP. Epigenetic impact of simulated maternal grooming on estrogen receptor alpha within the developing amygdala. Brain Behav Immun 2011; In press
- DonCarlos LL, Handa RJ. Developmental profile of estrogen receptor mRNA in the preoptic area of male and female neonatal rats. Brain Res Dev Brain Res 1994; 79:283 - 289
- Yamamoto Y, Carter CS, Cushing BS. Neonatal manipulation of oxytocin affects expression of estrogen receptor alpha. Neuroscience 2006; 137:157 - 164
- Yokosuka M, Okamura H, Hayashi S. Postnatal development and sex difference in neurons containing estrogen receptor-alpha immunoreactivity in the preoptic brain, the diencephalon and the amygdala in the rat. J Comp Neurol 1997; 389:81 - 93
- Auger CJ, Coss D, Auger AP, Forbes-Lorman RM. Epigenetic control of vasopressin expression is maintained by steroid hormones in the adult male rat brain. Proc Natl Acad Sci USA 2011; 108:4242 - 4247
- Hendrich B, Bird A. Identification and characterization of a family of mammalian methyl-CpG binding proteins. Mol Cell Biol 1998; 18:6538 - 6547
- Meehan RR, Lewis JD, McKay S, Kleiner EL, Bird AP. Identification of a mammalian protein that binds specifically to DNA containing methylated CpGs. Cell 1989; 58:499 - 507
- Amir RE, Van den Veyver IB, Wan M, Tran CQ, Francke U, Zoghbi HY. Rett syndrome is caused by mutations in X-linked MECP2, encoding methyl-CpG-binding protein 2. Nat Genet 1999; 23:185 - 188
- Chen RZ, Akbarian S, Tudor M, Jaenisch R. Deficiency of methyl-CpG binding protein-2 in CNS neurons results in a Rett-like phenotype in mice. Nat Genet 2001; 27:327 - 331
- Guy J, Hendrich B, Holmes M, Martin JE, Bird A. A mouse Mecp2-null mutation causes neurological symptoms that mimic Rett syndrome. Nat Genet 2001; 27:322 - 326
- Guy J, Gan J, Selfridge J, Cobb S, Bird A. Reversal of neurological defects in a mouse model of Rett syndrome. Science 2007; 315:1143 - 1147
- Nagarajan RP, Hogart AR, Gwye Y, Martin MR, LaSalle JM. Reduced MeCP2 expression is frequent in autism frontal cortex and correlates with aberrant MECP2 promoter methylation. Epigenetics 2006; 1:1 - 11
- Kurian JR, Forbes-Lorman RM, Auger AP. Sex difference in mecp2 expression during a critical period of rat brain development. Epigenetics 2007; 2:173 - 178
- Kurian JR, Bychowski ME, Forbes-Lorman RM, Auger CJ, Auger AP. Mecp2 organizes juvenile social behavior in a sex-specific manner. J Neurosci 2008; 28:7137 - 7142
- Klose RJ, Bird AP. Genomic DNA methylation: the mark and its mediators. Trends Biochem Sci 2006; 31:89 - 97
- Cunliffe VT. Eloquent silence: developmental functions of Class I histone deacetylases. Curr Opin Genet Dev 2008; 18:404 - 410
- Horlein AJ, et al. Ligand-independent repression by the thyroid hormone receptor mediated by a nuclear receptor co-repressor. Nature 1995; 377:397 - 404
- Cheng S, Brzostek S, Lee SR, Hollenberg AN, Balk SP. Inhibition of the dihydrotestosterone-activated androgen receptor by nuclear receptor corepressor. Mol Endocrinol 2002; 16:1492 - 1501
- Yoon HG, Wong J. The corepressors silencing mediator of retinoid and thyroid hormone receptor and nuclear receptor corepressor are involved in agonist- and antagonist-regulated transcription by androgen receptor. Mol Endocrinol 2006; 20:1048 - 1060
- Lavinsky RM, Jepsen K, Heinzel T, Torchia J, Mullen TM, Schiff R, et al. Diverse signaling pathways modulate nuclear receptor recruitment of N-CoR and SMRT complexes. Proc Natl Acad Sci USA 1998; 95:2920 - 2925
- Liu Z, Auboeuf D, Wong J, Chen JD, Tsai SY, Tsai MJ, et al. Coactivator/corepressor ratios modulate PR-mediated transcription by the selective receptor modulator RU486. Proc Natl Acad Sci USA 2002; 99:7940 - 7944
- Yoon HG, Chan DW, Reynolds AB, Qin J, Wong J. N-CoR mediates DNA methylation-dependent repression through a methyl CpG binding protein Kaiso. Mol Cell 2003; 12:723 - 734
- Cukier HN, Perez AM, Collins AL, Zhou Z, Zoghbi HY, Botas J. Genetic modifiers of MeCP2 function in Drosophila. PLoS Genet 2008; 4:1000179
- Kokura K, Kaul SC, Wadhwa R, Nomura T, Khan MM, Shinagawa T, et al. The Ski protein family is required for MeCP2-mediated transcriptional repression. J Biol Chem 2001; 276:34115 - 34121
- Alland L, Muhle R, Hou H Jr, Potes J, Chin L, Schreiber-Agus N, et al. Role for N-CoR and histone deacetylase in Sin3-mediated transcriptional repression. Nature 1997; 387:49 - 55
- Jessen HM, Kolodkin MH, Bychowski ME, Auger CJ, Auger AP. The nuclear receptor corepressor has organizational effects within the developing amygdala on juvenile social play and anxiety-like behavior. Endocrinology 2010; 151:1212 - 1220