Abstract
There is growing evidence the microbiota of the large bowel may influence the risk of developing colorectal cancer as well as other diseases including type-1 diabetes, inflammatory bowel diseases and irritable bowel syndrome. Current sampling methods to obtain microbial specimens, such as feces and mucosal biopsies, are inconvenient and unappealing to patients. Obtaining samples through rectal swabs could prove to be a quicker and relatively easier method, but it is unclear if swabs are an adequate substitute. We compared bacterial diversity and composition from rectal swabs and rectal mucosal biopsies in order to examine the viability of rectal swabs as an alternative to biopsies. Paired rectal swabs and mucosal biopsy samples were collected in un-prepped participants (n = 11) and microbial diversity was characterized by Terminal Restriction Fragment Length polymorphism (T-RFLP) analysis and quantitative polymerase chain reaction (qPCR) of the 16S rRNA gene. Microbial community composition from swab samples was different from rectal mucosal biopsies (p = 0.001). Overall the bacterial diversity was higher in swab samples than in biopsies as assessed by diversity indexes such as: richness (p = 0.01), evenness (p = 0.06) and Shannon’s diversity (p = 0.04). Analysis of specific bacterial groups by qPCR showed higher copy number of Lactobacillus (p < 0.0001) and Eubacteria (p = 0.0003) in swab samples compared with biopsies. Our findings suggest that rectal swabs and rectal mucosal samples provide different views of the microbiota in the large intestine.
Introduction
Increasing evidence suggests a role for the intestinal microbiota in colorectal cancer (CRC),Citation1 colorectal adenomasCitation2 and several other conditions such as inflammatory bowel diseases (ulcerative colitis and Crohn’s disease),Citation3 irritable bowel syndrome (IBS),Citation4 obesityCitation5 and type-1 diabetes.Citation6 The launch of the Human Microbiome ProjectCitation7 and the advent of molecular techniques have greatly increased our ability to identify and characterize microbial communities, thus improving our understanding of the role of the microbiota in common chronic diseases.
Currently, gut bacterial diversity in the human colon is determined through analysis of the luminal content (stool) and mucosal biopsies. Colorectal biopsies capture the diversity of flora in the mucosal layer of the large intestine where adherent bacteria reside.Citation8,Citation9 The bacteria in this compartment are of interest because of their direct interaction with the host immune system, and by consequence, their possible direct link to disease development.Citation10 Unfortunately, methods for obtaining colorectal biopsies such as sigmoidoscopy, anoscopy or colonoscopy are expensive and time consuming and may subject the patient to discomfort and inconveniences associated with the procedures.Citation11 Stool sampling, which does not pose a major risk to patients, is least liked because of the patient distaste for handling feces. A simpler, standardized, risk-free and inexpensive method to sample the gut bacteria would represent an important contribution.
In this study, we compared rectal swabs as a noninvasive low-risk sampling method and rectal mucosal biopsies obtained via unprepped, rigid sigmoidoscopy to assess the bacterial community composition and diversity of the human gut using terminal restriction fragment length polymorphism (T-RFLP) and quantitative PCR (qPCR) of the bacterial 16S rRNA gene. We hypothesized that rectal swabs have comparable bacterial diversity to rectal mucosal biopsies from the same participant.
Results
Study population
The mean age of participants was 56.3 y ± 5.6. Forty-five percent of the participants were male, and the average body mass index (BMI) was 30.5 ± 6.4 (). Rectal mucosal biopsies were obtained via rigid sigmodoscopy at approximately 10 cm from the anal verge while swabs were obtained 1–2 cm from the anal verge. Participants did not undergo colonic cleansing preparation prior to sample collection.
Table 1. Characteristic of study population (n = 11)
Analysis of T-RFLP profiles showed overall differences in community composition between swabs and biopsy samples
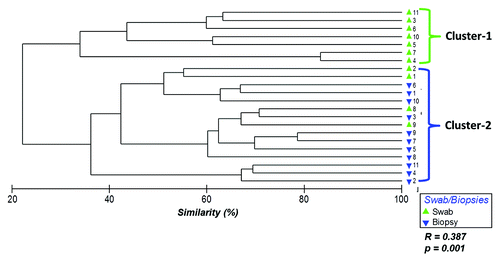
Hierarchical clustering of the 16S rRNA gene Terminal Restriction Fragments (T-RFs) based on Bray-Curtis similarities showed two main clusters suggesting differences in bacterial communities between samples collected from rectal swabs and biopsies (ANOSIM R = 0.387, p = 0.001) (). Cluster-1 was comprised entirely of rectal swab samples (100%) while cluster-2 was composed mainly of biopsy samples (73% biopsies and 27% swabs). Adenoma case/control analysis showed the results were independent of adenoma status. The cluster analysis for cases and controls are shown in Figure S1.
Figure 1. Hierarchical clustering of bacterial community profiles in rectal swabs and rectal biopsies. Bray-Curtis similarities were used to construct a dendrogram composed of the samples provided by the participants (1–11). Each participant is represented twice: rectal swab (green triangles) and rectal biopsy (blue triangles).
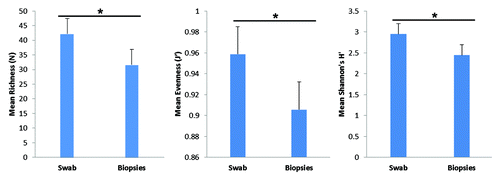
Using similarity percentage analysis (SIMPER), we assessed which specific T-RFs contributed to the differences between swabs and biopsies. A total of 26 T-RFs accounted for the overall diversity for the two groups, with a higher number of unique T-RFs in rectal swab samples than rectal biopsies (). Sixteen T-RFs were unique to swab samples (107, 108, 110, 112, 113, 146, 35, 387, 39, 399, 51, 53, 58, 59, 61, 62), while 2 TRFs (369 and 72) were unique to biopsy samples. Distribution of T-RFs for each individual sample as well as Bray-Curtis similarities matrix showed marked differences between swabs and biopsies from the same participant (Fig. S2A and B). We also assessed measures of microbial diversity namely richness (N), evenness (J’) and Shannon’s H (diversity) and observed that overall diversity measures were higher in rectal swabs compared with rectal biopsies (). Altogether, the T-RFLP results demonstrate that the bacterial community composition from rectal swabs and rectal biopsies are different.
Figure 2. A Distribution of terminal-restriction fragments (T-RFs) in rectal swabs and rectal biopsies. Bars represent the average abundance of each T-RF grouped by biopsies (red) or swabs (blue). Asterisks represent T-RFs that are significantly different (p < 0.05) between rectal biopsies and rectal swabs as assessed by t-test.
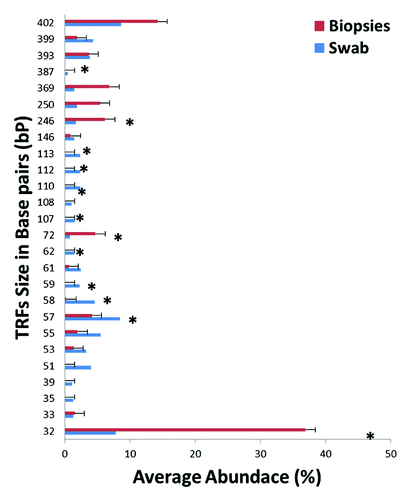
Figure 3. Measures of T-RF diversity in rectal swabs and rectal biopsies. Bars represent average diversity as estimated by T-RF richness (p = 0.014), evenness (p = 0.058) and Shannon’s diversity (p = 0.04). Calculated standard error is represented atop each bar graph. Statistical significance (*) was calculated by t-test.
Quantitative PCR showed differences in abundance of specific bacterial groups between swabs and biopsy samples
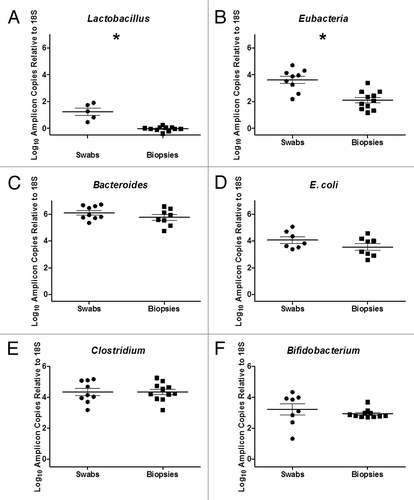
Clostridium spp, Bifidobacterium spp, Bacteroides spp, Lactobacillus spp and E. coli, bacteria groups that have been shown to be predominant members of the human gut microbiotaCitation1,Citation4,Citation12 were quantified by qPCR of the 16S rRNA gene and the results were normalized to human 18S RNA gene. All quantified bacterial groups and Eubacteria bacterial groups (as assessed by Universal 16S rRNA primers) showed higher abundance in swab specimens compared with biopsy samples. However, statistically significant differences were only observed for Lactobacillusspp and Eubacteria ().
Figure 4. Quantitative PCR of Bacterial 16S RNA Gene of (A) Lactobacillus spp, (B) Eubacteria, (C) Bacteroides spp., (D) E. coli, (E) Clostridium spp and (F) Bifidobacterium spp in rectal swabs and rectal biopsies. A significant increase in Lactobacillus spp (p < 0.0001) and Eubacterium spp (p = 0.0003) was observed in rectal swabs compared with rectal biopsies (*).
Discussion and Conclusions
The association between colorectal adenomas and dysbiosis of gut microbes has been previously reportedCitation2,Citation13 and could serve as the basis to identify microbial signatures that could lead to the development of tests to identify individuals at risk of developing colorectal cancer. Biopsies collected during colonoscopy, as well as stool samples, are the current methods to characterize the microbiota of the large intestine. A simple, standardized, risk-free and inexpensive method to assess bacterial community composition of the gut could lower the risks and inconvenience associated with collection of these samples. In the present study, we systematically compared the bacterial composition of rectal swabs and rectal mucosal biopsies collected during an un-prepped sigmoidoscopy from 11 participants. We assessed whether bacterial community composition from these two sampling sites is comparable and whether rectal swabs could be a viable alternative to currently used methods.
We used 16S rRNA gene T-RFLP fingerprinting analysis to reveal significant differences in the bacteria community profiles of samples collected via rectal swabs vs. mucosal biopsies. Similarly, bacterial diversity indexes showed significant differences between the two sampling sites. Swab samples had higher bacterial abundance and diversity compared with rectal mucosal biopsies. Durbán et al.Citation14 compared bacterial community composition of stool samples and rectal mucosal biopsies obtained from an un-prepped population of healthy participants. They reported that fecal and mucosal bacterial diversity from the same subject are different. In a study that compared healthy subjects to IBS subjects, Carroll et al.Citation4 observed reduced bacterial abundance and diversity in mucosal samples compared with stool samples from the same subjects. Our findings are compatible with the reports of Carroll et al. and Durbán et al. although we extended those findings to rectal swabs compared with biopsies. Similar to these and previous studies,Citation15 our results suggest that different niches within the large intestine possess distinct bacterial populations.
This is the first study we are aware of to compare gut microbial composition of samples collected via rectal swabs vs. rectal biopsies in subjects who had adenomas and those without adenomas. Additionally, investigating noninvasive alternatives for stratification of risk for colorectal cancer has the potential to increase screening rate and screening compliance among the population at risk since some participants may prefer to utilize easier and more convenient screening methods.Citation16,Citation17 Our findings from T-RFLP analysis showed statistically significant differences in the bacterial profiles from rectal swabs and mucosal biopsies. These results suggest that a quick fingerprinting technique could be efficiently used to compare bacterial community profiles before investing additional costs and time with more advanced sequencing technologies.
Our samples were obtained from un-prepped participants, which is a problem because it could increase the chances of contamination of rectal swabs with luminal content.Citation18 Since previous studies have observed that the luminal cavity and the colonic mucosa contain distinct bacterial communities,Citation14,Citation19-Citation21 use of un-prepped participants for sampling may have mixed those two bacterial communities. Another source of swab contamination may have been from local skin flora due to inadvertent swab contact with adjacent skin prior to insertion through the anus. We selected Staphylococcus aureus (S. aureus), a predominant skin microbe,Citation22 to assess the level of contamination of skin bacteria in swabs and biopsies. S. aureus was considerably more abundant in swabs (3.2 ± 0.192) than biopsies (1.6 ± 0.075) suggesting a potential contamination of swab samples with skin flora. In future studies, collecting samples through a sheath might reduce contamination by skin microbes. Finally, future studies should include a larger study population that samples several sites such as luminal, rectal swabs and biopsies in order to get a better picture of the microbial populations in the large intestine.
In summary, our findings suggest that the bacterial diversity in samples collected via rectal swabs and mucosal biopsies are different. While differences in bacterial community composition can be attributed to a whole array of factors, including host genetics and the environment, our sampling scheme enabled us to observe the diversity associated with two different sampling locations. Our results suggest potential differences in the niches within the human large intestine in relation to bacterial communities. Moreover, the differences in bacterial community composition that we observed suggest that both, swab sampling and biopsy collection, may be needed in order to get the full spectrum of the microbial community composition of the gut. Characterizing these unique bacterial communities of the large intestine is a first step toward understanding the complex association between bacterial diversity in the gut and intestine and disease development.
Materials and Methods
Study population and sampling
Study population included 11 participants enrolled as part of an ongoing curcumin trial at UNC Hospitals. Eligibility criteria included: good general health, age 40–80 y, willingness to follow the study protocol and provision of informed consent. As part of the study protocol, two swab samples were collected for each participant prior to sigmoidoscopy. Swab specimens were collected by inserting a sterile cotton-tipped swab (Cat. #14–959–82, Fisher Scientific) 1–2 cm beyond the anus and rotating for several seconds. Swabs were then placed into sterile phosphate buffered saline (PBS), vortexed for at least 2 min to ensure release of bacteria and stored at -80°C until further processing. Rectal mucosal biopsies were obtained through a rigid disposable sigmoidoscope (Welch Allyn KleenSpec Disposable Sigmoidoscope with Obturator) coated with gel and inserted to approximately 10 cm with the participant in the left lateral position. Disposable flexible biopsy forceps (Olympus EndoJaw Alligator Jaw-Step, Shinjuku) were used to obtain single mucosal pinches (3–10 mg) from two separate sites. Biopsy samples were rinsed in sterile PBS as previously described,Citation2,Citation13 snap-frozen, and then stored at -80°C until further processing. All samples for this study were collected prior to initiating treatment for all participants. Swab samples for two participants were excluded from qPCR analysis because of insufficient DNA. The study was approved by the Institutional Review Board (IRB) at the University of North Carolina School of Medicine (Protocol #10-1524)
DNA extractions and terminal restriction fragments length polymorphisms (T-RFLPs)
DNA extraction was performed on 500 µl of pelleted swab suspension and mucosal biopsy pinches (3–10 mg) using Qiagen’s DNeasy Blood and Tissue kit (Cat # 69504). Samples were treated with lysozyme followed by bead beating on a bullet blender homogenizer (Next Advance, Inc.), using a modified protocol.Citation8 T-RFLP is a PCR-based fingerprinting method to assess bacterial composition in gut samples. T-RFLP profiles were collected on both biopsy and swab samples following a previous protocol described by Shen et al. 2010.Citation2 For both biopsies and swabs, a standard amount of DNA (50 ng) was used as template for PCR. Swab samples for two participants were excluded from qPCR analysis because of insufficient DNA.
Quantitative PCR (qPCR) to assess specific bacteria known to be present in the human gut
Clostridium spp, Bifidobacteria spp, Bacteroides spp, and Lactobacillus spp and E. coli,Citation4 bacterial groups that have been shown to be predominant members of the human gut microbiota as described by previous studiesCitation1,Citation4,Citation12 were quantified using primers for PCR amplification of the 16S rRNA (rRNA) gene specific for each bacteria groups (Refer to Carroll et al.Citation4 for primer sequences). Also, universal 16S rRNA primers were used to capture all bacterial diversity for each sample henceforth referred as Eubacteria while Staphyloccocus aureus (S. aureus)Citation23 16S rRNA primers were used to assess contamination with skin bacteria. Modifications to the original protocol by Carroll et al.Citation4 included: the use of Fast SYBR Green Master Mix (Applied Biosystems, P/N: 4385614) and dilution of template DNA to a 1:10 (Clostridium, Bifidobacteria, Lactobacillus and Eubacteria) and 1:100 (Bifidobacteria, S. aureus and E. coli). Finally, the copy number for group-specific bacterial 16S rRNA gene was calculated based on a standard curve, which was adjusted to a starting DNA concentration of 50 ng/µL using the following formula to the unadjusted values: [50 ng/(A/B)] X unadjusted copy#
Here A is the concentration of the template DNA and B is the dilution factor; either 1:10 or 1:100. All bacterial specific quantifications were normalized to human 18S rRNA gene to account for the proportion of bacteria to human DNA in the samples. The ratio of bacterial-specific 16S abundance to human 18S rRNA abundanceCitation24 was used to determine the copy number and the differences between swabs and biopsies were compared by t-Test. Swab samples for two participants were excluded from qPCR analysis because of insufficient DNA leaving 9 swab samples for analysis.
Data analysis
We compared T-RFLP profiles from swabs and biopsies to determine bacterial community composition and diversity. The T-RF (phylotype) peaks size and area were determined by GeneMapper (Applied Biosystems Inc.). Peak area and fluorescence data were normalized and processed as described by Abdo et al.Citation25 The contribution of individual T-RFs was calculated as a proportion of the total T-RF peak area for each sample. For our analysis, we used these proportions rather than absolute numbers. The data matrix was used to generate Bray-Curtis similarities, a pairwise index of the degree of similarity, where values range from 0–100%. The generated values were then used to construct hierarchical clusters to observe grouping of samples base TRF abundance. We compared the similarities between groups (rectal swab/biopsy) by analysis of similarities (ANOSIM), a non-parametric test, were the significance is computed by permutation of group membership with 999 replicates. The test statistic R, which measures the strength of the correlations ranges from -1 to 1. An R value of 1 signifies differences between groups while an R value of 0 signifies that the groups are identical.
To determine the specific phylotypes that contributed to the differences in bacterial composition between swabs and biopsies we used similarity percentage (SIMPER) to compute the proportions of phylotypes for each group. Differences in bacterial richness (measure of the number of phylotypes) evenness (measure of how evenly the individuals are distributed among different phylotpes) and Shannon diversity index (measure of diversity) as well as mean bacterial 16S gene copy number between rectal swabs and biopsies were evaluated by t-test. The data analysis protocol has been previously described Shen et al. 2010Citation2 and was performed with the Primer 6 statistical package (PRIMER E).
| Abbreviations: | ||
| T-RFLP | = | terminal restriction fragment length polymorphism |
| qPCR | = | quantitative polymerase chain reaction |
| CRC | = | colorectal cancer |
| IBS | = | Irritable Bowel Syndrome |
| BMI | = | body mass index |
| SIMPER | = | similarity percentage |
| PBS | = | phosphate buffered saline |
Additional material
Download Zip (250.3 KB)Disclosure of Potential Conflicts of Interest
The authors disclose no conflict.
Acknowledgments
Funding for this study was provided by award number UL1RR025747 from the NIH National Center for Research Resources and P50CA106991 from the NIH National Cancer Institute. R01 CA136887, R01CA44684, P50CA106991, Center for Gastrointestinal Biology and Disease (CGIBD) P30 DK 034987.
Supplemental Materials
Supplemental materials may be found here:
http://www.landesbioscience.com/journals/gutmicrobes/article/22157/
References
- Sobhani I, Tap J, Roudot-Thoraval F, Roperch JP, Letulle S, Langella P, et al. Microbial dysbiosis in colorectal cancer (CRC) patients. PLoS One 2011; 6:e16393; http://dx.doi.org/10.1371/journal.pone.0016393; PMID: 21297998
- Shen XJ, Rawls JF, Randall T, Burcal L, Mpande CN, Jenkins N, et al. Molecular characterization of mucosal adherent bacteria and associations with colorectal adenomas. Gut Microbes 2010; 1:138 - 47; http://dx.doi.org/10.4161/gmic.1.3.12360; PMID: 20740058
- Gersemann M, Wehkamp J, Stange EF. Innate immune dysfunction in inflammatory bowel disease. J Intern Med 2012; 271:421 - 8; http://dx.doi.org/10.1111/j.1365-2796.2012.02515.x; PMID: 22324936
- Carroll IM, Chang YH, Park J, Sartor RB, Ringel Y. Luminal and mucosal-associated intestinal microbiota in patients with diarrhea-predominant irritable bowel syndrome. Gut Pathog 2010; 2:19; http://dx.doi.org/10.1186/1757-4749-2-19; PMID: 21143915
- Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature 2006; 444:1027 - 31; http://dx.doi.org/10.1038/nature05414; PMID: 17183312
- Brown CT, Davis-Richardson AG, Giongo A, Gano KA, Crabb DB, Mukherjee N, et al. Gut microbiome metagenomics analysis suggests a functional model for the development of autoimmunity for type 1 diabetes. PLoS One 2011; 6:e25792; http://dx.doi.org/10.1371/journal.pone.0025792; PMID: 22043294
- Turnbaugh PJ, Ley RE, Hamady M, Fraser-Liggett CM, Knight R, Gordon JI. The human microbiome project. Nature 2007; 449:804 - 10; http://dx.doi.org/10.1038/nature06244; PMID: 17943116
- Savage DC. Microbial ecology of the gastrointestinal tract. Annu Rev Microbiol 1977; 31:107 - 33; http://dx.doi.org/10.1146/annurev.mi.31.100177.000543; PMID: 334036
- Sonnenburg JL, Angenent LT, Gordon JI. Getting a grip on things: how do communities of bacterial symbionts become established in our intestine?. Nat Immunol 2004; 5:569 - 73; http://dx.doi.org/10.1038/ni1079; PMID: 15164016
- Goto Y, Kiyono H. Epithelial barrier: an interface for the cross-communication between gut flora and immune system. Immunol Rev 2012; 245:147 - 63; http://dx.doi.org/10.1111/j.1600-065X.2011.01078.x; PMID: 22168418
- n.d.) ACS. Colorectal Cancer Facts & Figures. In: Society AC, ed., 2011:1-30.
- Tap J, Mondot S, Levenez F, Pelletier E, Caron C, Furet JP, et al. Towards the human intestinal microbiota phylogenetic core. Environ Microbiol 2009; 11:2574 - 84; http://dx.doi.org/10.1111/j.1462-2920.2009.01982.x; PMID: 19601958
- Sanapareddy N, Legge RM, Jovov B, McCoy A, Burcal L, Araujo-Perez F, et al. Increased rectal microbial richness is associated with the presence of colorectal adenomas in humans. ISME J 2012; http://dx.doi.org/10.1038/ismej.2012.43; PMID: 22622349
- Durbán A, Abellán JJ, Jiménez-Hernández N, Ponce M, Ponce J, Sala T, et al. Assessing gut microbial diversity from feces and rectal mucosa. Microb Ecol 2011; 61:123 - 33; http://dx.doi.org/10.1007/s00248-010-9738-y; PMID: 20734040
- Hong PY, Croix JA, Greenberg E, Gaskins HR, Mackie RI. Pyrosequencing-based analysis of the mucosal microbiota in healthy individuals reveals ubiquitous bacterial groups and micro-heterogeneity. PLoS One 2011; 6:e25042; http://dx.doi.org/10.1371/journal.pone.0025042; PMID: 21966408
- DeBourcy AC, Lichtenberger S, Felton S, Butterfield KT, Ahnen DJ, Denberg TD. Community-based preferences for stool cards versus colonoscopy in colorectal cancer screening. J Gen Intern Med 2008; 23:169 - 74; http://dx.doi.org/10.1007/s11606-007-0480-1; PMID: 18157581
- Wolf RL, Basch CE, Brouse CH, Shmukler C, Shea S. Patient preferences and adherence to colorectal cancer screening in an urban population. Am J Public Health 2006; 96:809 - 11; http://dx.doi.org/10.2105/AJPH.2004.049684; PMID: 16571715
- Chen W, Liu F, Ling Z, Tong X, Xiang C. Human intestinal lumen and mucosa-associated microbiota in patients with colorectal cancer. PLoS One 2012; 7:e39743; http://dx.doi.org/10.1371/journal.pone.0039743; PMID: 22761885
- Eckburg PB, Bik EM, Bernstein CN, Purdom E, Dethlefsen L, Sargent M, et al. Diversity of the human intestinal microbial flora. Science 2005; 308:1635 - 8; http://dx.doi.org/10.1126/science.1110591; PMID: 15831718
- Lepage P, Seksik P, Sutren M, de la Cochetière MF, Jian R, Marteau P, et al. Biodiversity of the mucosa-associated microbiota is stable along the distal digestive tract in healthy individuals and patients with IBD. Inflamm Bowel Dis 2005; 11:473 - 80; http://dx.doi.org/10.1097/01.MIB.0000159662.62651.06; PMID: 15867587
- Zoetendal EG, von Wright A, Vilpponen-Salmela T, Ben-Amor K, Akkermans AD, de Vos WM. Mucosa-associated bacteria in the human gastrointestinal tract are uniformly distributed along the colon and differ from the community recovered from feces. Appl Environ Microbiol 2002; 68:3401 - 7; http://dx.doi.org/10.1128/AEM.68.7.3401-3407.2002; PMID: 12089021
- Paju S, Bernstein JM, Haase EM, Scannapieco FA. Molecular analysis of bacterial flora associated with chronically inflamed maxillary sinuses. J Med Microbiol 2003; 52:591 - 7; http://dx.doi.org/10.1099/jmm.0.05062-0; PMID: 12808082
- Seetaram SP, Liu R, Ramotar K, Harder CJ. Real-time PCR detection of MRSA. Spartan Biosciences: Application Notes 2008; AN0007 Ver. 1.2.
- Rawls JF, Mahowald MA, Goodman AL, Trent CM, Gordon JI. In vivo imaging and genetic analysis link bacterial motility and symbiosis in the zebrafish gut. Proc Natl Acad Sci U S A 2007; 104:7622 - 7; http://dx.doi.org/10.1073/pnas.0702386104; PMID: 17456593
- Abdo Z, Schüette UM, Bent SJ, Williams CJ, Forney LJ, Joyce P. Statistical methods for characterizing diversity of microbial communities by analysis of terminal restriction fragment length polymorphisms of 16S rRNA genes. Environ Microbiol 2006; 8:929 - 38; http://dx.doi.org/10.1111/j.1462-2920.2005.00959.x; PMID: 16623749