Abstract
Faecalibacterium prausnitzii is a major commensal bacterium, and its prevalence is often decreased in conditions of intestinal dysbiosis. The phylogenic identity of this bacterium was described only recently. It is still poorly characterized, and its specific growth requirements in the human gastrointestinal tract are not known. In this review, we consider F. prausnitzii metabolism, its ecophysiology in both humans and animals, and the effects of drugs and nutrition on its population. We list important questions about this beneficial and ubiquitous commensal bacterium that it would be valuable to answer.
Keywords: :
Introduction
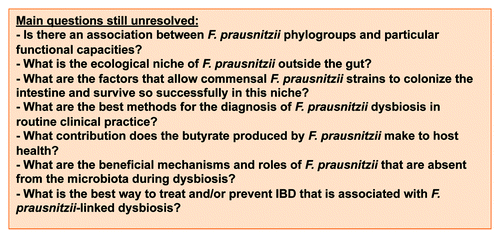
Faecalibacterium prausnitzii is a member of the phylum Firmicutes and a major component of human microbiota, but was first described only recently.Citation1 It has been the subject of few studies, partly because it is an extremely oxygen-sensitive (EOS) bacterium.Citation1 It is an atypical bacterium that has been difficult to classify in the bacterial nomenclature.Citation1 Analysis of the F. prausnitzii membrane suggests that this bacterium either lacks cell wall lipopolysaccharides (LPS) or displays an unusual LPS composition.Citation2 Over the last ten years, there has been substantial interest in F. prausnitzii in the microbiota of patients with intestinal and metabolic disorders, and particularly Inflammatory Bowel Disease (IBD) patients. These diseases are characterized by a dysbiosis, or in other words microbial imbalance (between “symbionts” and “pathobionts”), in the gut.Citation3 The Firmicutes-Bacteroidetes ratio is commonly affected with a decrease of F. prausnitzii population in such patients.Citation4 Recent studies report an association between low F. prausnitzii population levels and the risk of relapse in IBD. In ulcerative colitis (UC) patients, there is a clear correlation between F. prausnitzii population level and maintenance of clinical remission.Citation5 Similarly, in Crohn disease (CD) patients, a low relative count of this bacterium is risk factor for endoscopic recurrence within 6 months.Citation6 Interestingly, F. prausnitzii has immunomodulatory properties and is now considered as both an indicator of, and an actor in, human health in adults.Citation7 Although there have been various suggestions for the mechanisms involved, the role of F. prausnitzii in host immune responses is poorly understood. Human F. prausnitzii strains have been classified into two different molecular phylogroups, but no functional specificities have been linked to these phylogroups ().Citation8 Genomic data generated by microbiota metagenome projects will undoubtedly improve our knowledge of non-cultivable and difficult to cultivate strains. It may also be very informative to study the anti-inflammatory activities and molecular phylogroups of strains isolated from IBD patients and compare them to those of strains isolated from healthy individuals.
The role of F. prausnitzii in the homeostasis of the crosstalk between host and microbiota is unlikely to be restricted to its anti-inflammatory potential.Citation6 Indeed, the biological effects of F. prausnitzii may be also linked to its localization in the gastrointestinal tract (GIT), its metabolic activities, and its complementarities with other bacteria of the microbiota. In this review, we consider where and when F. prausnitzii may affect host physiology. Various unresolved questions that we believe important are listed in . We also propose an approach to develop a novel personalized treatment strategy based on using medicine and nutrition to modulate the F. prausnitzii population.
F. prausnitzii: A Late but Major Commensal Colonizer of the GIT
F. prausnitzii is usually described as an EOS bacterium but is able to grow in micro-aerobic conditions by using extracellular electron transfer in the presence of flavins and cysteine or glutathione.Citation1,Citation9 This capacity may explain how such anaerobic bacteria could colonize niches, including the gut mucosa, where there is an oxygen gradient.Citation9,Citation10 Nevertheless, it is difficult to cultivate F. prausnitzii, and various molecular approaches have been used to evaluate F. prausnitzii populations: (1) detection of 16S rRNA gene sequences,Citation11,Citation12 (2) PCR techniques based on single primers,Citation13,Citation14 (3) assaying 16S RNA by membrane-array methodsCitation15 or hybridization techniques, and (4) in situ hybridization.Citation16,Citation17F. prausnitzii DNA was found in the recently described metagenome catalog and is now considered to be a major member of the phylogenetic core.Citation18,Citation19 The abundance and ubiquity of F. prausnitzii suggest that it is a major contributor to microbiota functions in healthy individuals. It is therefore important to determine both the kinetics of its implantation and its localization in the GIT.
Temporal colonization in humans ()
Figure 2. Kinetics of implantation of F. prausnitzii. Changes in human fecal F. prausnitzii populations with host age (adapted from Hopkins et al., Balamurugan et al. and Van Tongeren et al.).Citation21,Citation22,Citation25
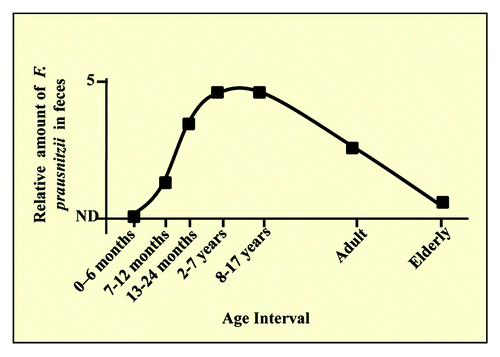
Although F. prausnitzii is dominant in healthy adults, its population in the intestine is modulated by diverse factors. A recent study suggests that the amount of F. prausnitzii in the gut microbiota depends on the sex of the host: there is less in human females than males (female to male ratio: 0.41, P ≤ 0.05 as evaluated from gut microbiota DNA).Citation20 Several reports indicate that the populations of this bacterium change with age. The amount of F. prausnitzii-specific RNA in stools from babies up to the age of 6 months is below the detection threshold; the value then increases between ages 6 and 24 months but remains low until early childhood (2–3 years).Citation21-Citation24 In elderly persons, there is a significant decrease of F. prausnitzii to 0.3%.Citation25 The low F. prausnitzii populations in early infancy suggest that the arrival of initial colonizers may facilitate subsequent implantation of F. prausnitzii. Possibly, consumption of the available oxygen by facultative anaerobic bacteria is required to generate an anaerobic environment favorable for the growth of obligatory anaerobic bacteria such as F. prausnitzii.Citation26 The implantation of EOS bacteria and specifically F. prausnitzii depends on the physico-chemical conditions previously created by other commensal bacteria.Citation27 Rezzonico et al. found that after inoculation of germ-free mice with a simplified human microbiota, all tested strains were systematically detected in all animals except for the reference strain of F. prausnitzii A2–165 (DSM17677).Citation28 In these experiments, all bacteria were introduced at the same time, and this did not allow efficient implantation of F. prausnitzii. A recent study describes F. prausnitzii in mono-colonized recipient germ-free mice,Citation29 but we have been unable to obtain rats mono-colonized by F. prausnitzii: prior colonization by Bacteroides thetaiotaomicron was required for robust implantation of F. prausnitzii in a rat model.Citation30 After 4 weeks of preparation of the GIT by B. thetaiotaomicron, a stable balance was maintained between the two bacteria, with B. thetaiotaomicron counts remaining 100-fold higher than F. prausnitzii counts.Citation30 These investigations with various rodent models maintained in germ-free conditions suggested that oxygen tension is an important determinant of colonization of the gut by F. prausnitzii. According to the “oxygen hypothesis” proposed by Rigottier-Gois,Citation31 oxygen is a major factor shaping patterns of colonization by EOS gut microbes. These observations provide insights into mechanisms governing microbial ecology and processes of colonization; they also raise questions about the ecological niche of the various strains outside the GIT ().
Colonization along the human GIT
F. prausnitzii implantation varies along the GIT, with a significantly higher population in the proximal colon than in the terminal ileum,Citation32 and few differences have been observed between the numbers of this organism in different parts of the large bowel. Relatively little is known about the interaction of F. prausnitzii with the mucus layer produced by the intestinal epithelium. Because of its distribution in the GIT, F. prausnitzii has been called a “fecomucus” bacterium: the highest concentration is in feces, and it is less abundant but detectable in mucus.Citation33,Citation34F. prausnitzii can survive in the adjacent mucosa where there is an oxygen influx from the gut epithelium. Inside the gut, its growth and survival (at the oxygenated fecal-mucosal interface) seems to depend on extracellular redox mediators such as flavin.Citation9,Citation10 Thus, the distribution of F. prausnitzii along the longitudinal and luminal axes of the gut are determined by a combination of several environmental factors, including the distribution of redox mediators, oxygen concentration, other bacteria, the mucus layer, bile salt concentrations, and pH.Citation8,Citation35 In rats, F. prausnitzii implantation is in part dependent on the same factors and it contributes to intestinal homeostasis mainly through effects on cell differentiation, and especially that of cells of the secretory lineage.Citation30 A better understanding is required of the environmental factors allowing the survival and the growth of F. prausnitzii in the gut ().
F. prausnitzii in animal intestinal microbiota
F. prausnitzii is widely distributed in the GIT of mammals. Interestingly, in pigs, FISH analyses showed that the localization of F. prausnitzii-related bacteria is very similar to that in humans. It is abundant in the hind gut (proximal colon 2 ± 0.5% and rectum 2.4 ± 0.7% of dominant bacteria) but was below the detection threshold in both the stomach and jejunum.Citation36F. prausnitzii has been detected in the microbiota of pigs and piglets,Citation37,Citation38 calves,Citation39 poultry including chickens and turkeys,Citation40-Citation46 and mice.Citation47 Most of these strains share less than 97% sequence identity with the human strain in the 16S rRNA gene and are thus named F. prausnitzii-like strains.Citation36,Citation40-Citation42 They are predominant bacteria in the intestines of many mammals and also in some insects. Indeed, under its initial name of Fusobacterium prausnitzii, F. prausnitzii has been found in the hind gut of the cockroach Eublaberus posticus.Citation48 These descriptions have led some authors to suggest that each animal species has its own distinctive set of phylotypes related to F. prausnitzii in its GIT.Citation41
What is Known about F. prausnitzii Metabolism?
Human F. prausnitzii has been considered to be “a key functional member of the core microbiome that most influences host metabolism and hence health”.Citation49 This role is in part due to F. prausnitzii being one of the most abundant of the butyrate-producing bacteria in the GIT. However, it is not known whether it is the major butyrate producer of the intestinal microbiota. Butyrate is a short chain fatty acid (SCFA) and very important in gut physiology and in the systemic functions and beneficial effects of the gut microbiota for human health.Citation50 Analysis of SCFAs pattern in stools from CD patients shows higher than normal proportions of acetate (70%) and low proportions of propionate and butyrate (14.9% and 7.99%, respectively).Citation51 However, it is not yet clear whether the production of butyrate by F. prausnitzii is directly linked to host responsiveness or health benefits. It would be informative to construct F. prausnitzii mutants defective for butyrate synthesis and use them to evaluate the effect of butyrate produced directly in situ by F. prausnitzii (). The metabolic activity of F. prausnitzii is not restricted to the production of butyrate, and its potential for immunomodulation is also linked to other molecules and/or metabolites, but they have not yet been characterized ().Citation6
The production of SCFA by F. prausnitzii was described in vitro for the first time by using a complex rumen fluid-based medium in strict anaerobic conditions.Citation52 F. prausnitzii is an acetate consumer and butyrate producer, and it can also produce carbon dioxide, formate, and D-lactate, although none of the strains isolated to date produce hydrogen.Citation1,Citation53 In batch cultures, most of the carbon in the butyrate produced (around 85%) is derived from external acetate, with only 15% provided directly from glucose.Citation54 In 2002, Duncan et al.Citation55 detected a Butyryl CoA:acetate CoA transferase in the F. prausnitzii reference strain A2-165 in which no butyrate kinase activity was found. In the human GIT, F. prausnitzii produces butyrate associated with a consumption of both acetate and carbohydrates.Citation52,Citation54 Moreover, F. prausnitzii strains can hydrolyze fructose, fructo-oligosaccharide, apple pectin, and starch, and some can hydrolyze inulin.Citation1,Citation8,Citation56 None of the strains isolated to date are able to exploit as sole energy source any of arabinose, melibiose, raffinose, rhamnose, ribose, xylose, linear and α-1,2-branched dextrans, arabinogalactan, xylan, citrus pectin, or peptides.Citation1,Citation8,Citation57 Most F. prausnitzii strains can grow on the host-derived sugar N-acetylglucosamine and some strains on D-glucosamine and D-glucuronic acid; B-glucuronidase activity has been reported in some F. prausnitzii isolates.Citation8,Citation58 This suggests that F. prausnitzii is able to switch from diet- to host-derived substrates, a feature common to several major bacterial species in the human colon.Citation59,Citation60 No evidence has been found of porcine gastric mucin fermentation by F. prausnitzii.Citation8 No minimal medium has yet been described for F. prausnitzii growth although some strains are able to grow on simplified medium containing acetate.Citation1 The analysis of the metabolomic profiles of a large collection of strains isolated from both healthy subjects and patients suffering disease-associated dysbiosis would be very useful, in particular to document the metabolic activity of F. prausnitzii.
How Medicines and Nutrition May Modulate F. prausnitzii Population and Activity
Various treatments used for IBD patients, such as rifaximin,Citation61 interferon-α-2b,Citation62 cortisol, and infliximab,Citation33 have been shown to have a positive effect on the F. prausnitzii population in the microbiota. However, there is published evidence that a large number of xenobiotics may decrease the F. prausnitzii population in the microbiota. Antibiotic therapy, chemotherapy, isoflavones, and essential oils markedly decrease the richness of species of the Clostridium cluster IV and significantly reduce F. prausnitzii populations.Citation63-Citation66
The metabolism of colonic bacteria depends largely on fibers that are not digested by human enzymes in the upper GIT. Work with fiber-free and fiber-supplemented liquid diets found that F. prausnitzii populations and fecal butyrate correlate with the fiber input.Citation67 In vitro conditions mimicking those of the proximal colon show that high levels of dietary fiber significantly increased clostridial cluster XIVa and F. prausnitzii populations.Citation68 Other specific diets, like a raffinose diet, a chickpea diet, and a novel diet based on fibers such as polydextrose and soluble corn fiber, can increase F. prausnitzii abundance.Citation69,Citation70 Diet may affect F. prausnitzii populations directly or indirectly by enhancing metabolite cross-feeding between microbes. The benefits of fiber intake have been demonstrated in a murine model of IBD, and this work also suggested a link between fiber and F. prausnitzii levels.Citation71 However, elemental diet therapy (nutrients in an easily assimilated form essentially composed of amino acids, fats, sugars, vitamins, and minerals), used mainly in the treatment of CD patients may decrease fecal F. prausnitzii counts.Citation72 In fact, this type of diet permits only very small amounts of undigested food residues, and such residues are required for normal levels of microorganisms in the lower gut.
The effects of prebiotics, such as inulin, on bifidogenic and butyrogenic bacteria are well established. The inclusion of inulin-type fructans in the diet of obese women may affect the gut microbiota, including increases in the populations of F. prausnitzii species, and thereby may have a significant impact on several key metabolites involved in obesity and/or diabetes.Citation73 The intake of 10 g/day inulin over a 16-day period resulted in specific and significant modifications of the composition of the human microbiota characterized by an increase in both Bifidobacterium and F. prausnitzii.Citation74,Citation75 Moreover, in vitro experiments showed that some exopolysaccharides produced by Bifidobacterium pseudocatenulatum, a human intestinal strain of Bifidobacteria, could increase the prevalence of F. prausnitzii.Citation76 Similarly, a human study showed that B. longum BB536 intake (13 weeks treatment) enhanced F. prausnitzii 16S rRNA gene copy numbers in Japanese individuals with cedar pollinosis.Citation77 This is consistent with a putative symbiotic cooperation or cross feeding between F. prausnitzii and microbes generally recognized as beneficial, such as Bifidobacterium and Lactobacillus spp.Citation45,Citation46 For instance, Bifidobacteria are acetate producers in the gut, and one possible approach to increase the F. prausnitzii population is to feed Bifidobacteria, which then feed F. prausnitzii by producing acetate. However, the effects of probiotics are strain-specific. Indeed, a recent study has demonstrated that the intake of Lactobacillus johnsonii strain La1 by healthy volunteers decreased F. prausnitzii levels.Citation78
The consumption of some prebiotics or probiotics could enhance the concentrations of beneficial species and especially F. prausnitzii in the GIT. This type of approach is promising for patients with intestinal disorders, although relevant clinical trials performed to date included only small numbers of subjects and lack statistical power. We believe that it is likely that therapeutic strategies will need to be individually adapted to the findings of microbiota analysis, as proposed by SwidsinskiCitation33 ().
Conclusion
F. prausnitzii is a commensal bacterium; it is a major member of adult human microbiota and is also found in most animals. The time course of F. prausnitzii colonization has been described, but many questions about the specificity of the conditions required for its implantation have not been answered. The ubiquity and population level of F. prausnitzii and its frequent involvement in dysbiosis indicate that this bacterium is a major contributor to the functions of the microbiota and intestinal health. Modulation of F. prausnitzii populations may be useful for preventive or therapeutic treatments. However, it is still not clear how to treat and/or prevent IBD associated with F. prausnitzii dysbiosis, and it may be necessary to establish a personal diagnosis for each patient, based on microbiota analysis, to allow appropriate management (). Treatments complementary to standard therapy should be investigated, involving, for example, various nutritional strategies or prebiotics or probiotics that favor F. prausnitzii population expansion. Further research, and in particular work to elucidate the mutualistic interactions between F. prausnitzii and the host, may lead to valuable medical applications ().
Disclosure of Potential Conflicts of Interest
No potential conflict of interest was disclosed.
Acknowledgments
We thank Sylvie Hudault and Jean-Marc Chatel for fruitful discussions and critical reading of the manuscript. This review was a part of the FPARIS collaborative project selected and supported by the Vitagora Competitive Cluster and funded by the French FUI (Fond Unique Interministériel; FUI: n°F1010012D), the FEDER (Fonds Européen de Développement Régional; Bourgogne: 34606), the Burgundy Region, the Conseil Général 21, and the Grand Dijon. This work was also supported by Merck Médication Familiale (Dijon, France) and Biovitis (Saint Etienne de Chomeil, France). R.M. and S.M. each receive a salary from the same grants.
References
- Duncan SH, Hold GL, Harmsen HJ, Stewart CS, Flint HJ. Growth requirements and fermentation products of Fusobacterium prausnitzii, and a proposal to reclassify it as Faecalibacterium prausnitzii gen. nov., comb. nov. Int J Syst Evol Microbiol 2002; 52:2141 - 6; http://dx.doi.org/10.1099/ijs.0.02241-0; PMID: 12508881
- Jantzen E, Hofstad T. Fatty acids of Fusobacterium species: taxonomic implications. J Gen Microbiol 1981; 123:163 - 71; PMID: 7320695
- Round JL, Mazmanian SK. The gut microbiota shapes intestinal immune responses during health and disease. Nat Rev Immunol 2009; 9:313 - 23; http://dx.doi.org/10.1038/nri2515; PMID: 19343057
- Neish AS. Microbes in gastrointestinal health and disease. Gastroenterology 2009; 136:65 - 80; http://dx.doi.org/10.1053/j.gastro.2008.10.080; PMID: 19026645
- Varela E, Manichanh C, Gallart M, Torrejón A, Borruel N, Casellas F, Guarner F, Antolin M. Colonisation by Faecalibacterium prausnitzii and maintenance of clinical remission in patients with ulcerative colitis. Aliment Pharmacol Ther 2013; 38:151 - 61; http://dx.doi.org/10.1111/apt.12365; PMID: 23725320
- Sokol H, Pigneur B, Watterlot L, Lakhdari O, Bermúdez-Humarán LG, Gratadoux JJ, Blugeon S, Bridonneau C, Furet JP, Corthier G, et al. Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc Natl Acad Sci U S A 2008; 105:16731 - 6; http://dx.doi.org/10.1073/pnas.0804812105; PMID: 18936492
- Miquel S, Martín R, Rossi O, Bermúdez-Humarán LG, Chatel JM, Sokol H, Thomas M, Wells JM, Langella P. Faecalibacterium prausnitzii and human intestinal health. Curr Opin Microbiol 2013; 16:255 - 61; http://dx.doi.org/10.1016/j.mib.2013.06.003; PMID: 23831042
- Lopez-Siles M, Khan TM, Duncan SH, Harmsen HJ, Garcia-Gil LJ, Flint HJ. Cultured representatives of two major phylogroups of human colonic Faecalibacterium prausnitzii can utilize pectin, uronic acids, and host-derived substrates for growth. Appl Environ Microbiol 2012; 78:420 - 8; http://dx.doi.org/10.1128/AEM.06858-11; PMID: 22101049
- Khan MT, Duncan SH, Stams AJ, van Dijl JM, Flint HJ, Harmsen HJ. The gut anaerobe Faecalibacterium prausnitzii uses an extracellular electron shuttle to grow at oxic-anoxic interphases. ISME J 2012; 6:1578 - 85; http://dx.doi.org/10.1038/ismej.2012.5; PMID: 22357539
- Khan MT, Browne WR, van Dijl JM, Harmsen HJ. How can Faecalibacterium prausnitzii employ riboflavin for extracellular electron transfer?. Antioxid Redox Signal 2012; 17:1433 - 40; http://dx.doi.org/10.1089/ars.2012.4701; PMID: 22607129
- Rigottier-Gois L, Bourhis AG, Gramet G, Rochet V, Doré J. Fluorescent hybridisation combined with flow cytometry and hybridisation of total RNA to analyse the composition of microbial communities in human faeces using 16S rRNA probes. FEMS Microbiol Ecol 2003; 43:237 - 45; http://dx.doi.org/10.1111/j.1574-6941.2003.tb01063.x; PMID: 19719684
- Suau A, Bonnet R, Sutren M, Godon JJ, Gibson GR, Collins MD, Doré J. Direct analysis of genes encoding 16S rRNA from complex communities reveals many novel molecular species within the human gut. Appl Environ Microbiol 1999; 65:4799 - 807; PMID: 10543789
- Rochet V, Rigottier-Gois L, Rabot S, Doré J. Validation of fluorescent in situ hybridization combined with flow cytometry for assessing interindividual variation in the composition of human fecal microflora during long-term storage of samples. J Microbiol Methods 2004; 59:263 - 70; http://dx.doi.org/10.1016/j.mimet.2004.07.012; PMID: 15369862
- Wang RF, Cao WW, Cerniglia CE. PCR detection and quantitation of predominant anaerobic bacteria in human and animal fecal samples. Appl Environ Microbiol 1996; 62:1242 - 7; PMID: 8919784
- Wang RF, Beggs ML, Robertson LH, Cerniglia CE. Design and evaluation of oligonucleotide-microarray method for the detection of human intestinal bacteria in fecal samples. FEMS Microbiol Lett 2002; 213:175 - 82; http://dx.doi.org/10.1111/j.1574-6968.2002.tb11302.x; PMID: 12167534
- Hold GL, Schwiertz A, Aminov RI, Blaut M, Flint HJ. Oligonucleotide probes that detect quantitatively significant groups of butyrate-producing bacteria in human feces. Appl Environ Microbiol 2003; 69:4320 - 4; http://dx.doi.org/10.1128/AEM.69.7.4320-4324.2003; PMID: 12839823
- Suau A, Rochet V, Sghir A, Gramet G, Brewaeys S, Sutren M, Rigottier-Gois L, Doré J. Fusobacterium prausnitzii and related species represent a dominant group within the human fecal flora. Syst Appl Microbiol 2001; 24:139 - 45; http://dx.doi.org/10.1078/0723-2020-00015; PMID: 11403393
- Tap J, Mondot S, Levenez F, Pelletier E, Caron C, Furet JP, Ugarte E, Muñoz-Tamayo R, Paslier DL, Nalin R, et al. Towards the human intestinal microbiota phylogenetic core. Environ Microbiol 2009; 11:2574 - 84; http://dx.doi.org/10.1111/j.1462-2920.2009.01982.x; PMID: 19601958
- Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, Manichanh C, Nielsen T, Pons N, Levenez F, Yamada T, et al, MetaHIT Consortium. A human gut microbial gene catalogue established by metagenomic sequencing. Nature 2010; 464:59 - 65; http://dx.doi.org/10.1038/nature08821; PMID: 20203603
- Aguirre de Cárcer D, Cuív PO, Wang T, Kang S, Worthley D, Whitehall V, Gordon I, McSweeney C, Leggett B, Morrison M. Numerical ecology validates a biogeographical distribution and gender-based effect on mucosa-associated bacteria along the human colon. ISME J 2011; 5:801 - 9; http://dx.doi.org/10.1038/ismej.2010.177; PMID: 21124491
- Hopkins MJ, Macfarlane GT, Furrie E, Fite A, Macfarlane S. Characterisation of intestinal bacteria in infant stools using real-time PCR and northern hybridisation analyses. FEMS Microbiol Ecol 2005; 54:77 - 85; http://dx.doi.org/10.1016/j.femsec.2005.03.001; PMID: 16329974
- Balamurugan R, Janardhan HP, George S, Chittaranjan SP, Ramakrishna BS. Bacterial succession in the colon during childhood and adolescence: molecular studies in a southern Indian village. Am J Clin Nutr 2008; 88:1643 - 7; http://dx.doi.org/10.3945/ajcn.2008.26511; PMID: 19064526
- Wang M, Ahrné S, Antonsson M, Molin G. T-RFLP combined with principal component analysis and 16S rRNA gene sequencing: an effective strategy for comparison of fecal microbiota in infants of different ages. J Microbiol Methods 2004; 59:53 - 69; http://dx.doi.org/10.1016/j.mimet.2004.06.002; PMID: 15325753
- Fallani M, Rigottier-Gois L, Aguilera M, Bridonneau C, Collignon A, Edwards CA, Corthier G, Doré J. Clostridium difficile and Clostridium perfringens species detected in infant faecal microbiota using 16S rRNA targeted probes. J Microbiol Methods 2006; 67:150 - 61; http://dx.doi.org/10.1016/j.mimet.2006.03.010; PMID: 16647148
- van Tongeren SP, Slaets JP, Harmsen HJ, Welling GW. Fecal microbiota composition and frailty. Appl Environ Microbiol 2005; 71:6438 - 42; http://dx.doi.org/10.1128/AEM.71.10.6438-6442.2005; PMID: 16204576
- Adlerberth I, Wold AE. Establishment of the gut microbiota in Western infants. Acta Paediatr 2009; 98:229 - 38; http://dx.doi.org/10.1111/j.1651-2227.2008.01060.x; PMID: 19143664
- Tomas J, Wrzosek L, Bouznad N, Bouet S, Mayeur C, Noordine ML, Honvo-Houeto E, Langella P, Thomas M, Cherbuy C. Primocolonization is associated with colonic epithelial maturation during conventionalization. FASEB J 2013; 27:645 - 55; http://dx.doi.org/10.1096/fj.12-216861; PMID: 23118025
- Rezzonico E, Mestdagh R, Delley M, Combremont S, Dumas ME, Holmes E, Nicholson J, Bibiloni R. Bacterial adaptation to the gut environment favors successful colonization: microbial and metabonomic characterization of a simplified microbiota mouse model. Gut Microbes 2011; 2:307 - 18; http://dx.doi.org/10.4161/gmic.18754; PMID: 22157236
- Atarashi K, Tanoue T, Oshima K, Suda W, Nagano Y, Nishikawa H, Fukuda S, Saito T, Narushima S, Hase K, et al. Treg induction by a rationally selected mixture of Clostridia strains from the human microbiota. Nature 2013; 500:232 - 6; http://dx.doi.org/10.1038/nature12331; PMID: 23842501
- Wrzosek L, Miquel S, Noordine ML, Bouet S, Joncquel Chevalier-Curt M, Robert V, Philippe C, Bridonneau C, Cherbuy C, Robbe-Masselot C, et al. Bacteroides thetaiotaomicron and Faecalibacterium prausnitzii influence the production of mucus glycans and the development of goblet cells in the colonic epithelium of a gnotobiotic model rodent. BMC Biol 2013; 11:61; http://dx.doi.org/10.1186/1741-7007-11-61; PMID: 23692866
- Rigottier-Gois L. Dysbiosis in inflammatory bowel diseases: the oxygen hypothesis. ISME J 2013; 7:1256 - 61; http://dx.doi.org/10.1038/ismej.2013.80; PMID: 23677008
- Ahmed S, Macfarlane GT, Fite A, McBain AJ, Gilbert P, Macfarlane S. Mucosa-associated bacterial diversity in relation to human terminal ileum and colonic biopsy samples. Appl Environ Microbiol 2007; 73:7435 - 42; http://dx.doi.org/10.1128/AEM.01143-07; PMID: 17890331
- Swidsinski A, Loening-Baucke V, Vaneechoutte M, Doerffel Y. Active Crohn’s disease and ulcerative colitis can be specifically diagnosed and monitored based on the biostructure of the fecal flora. Inflamm Bowel Dis 2008; 14:147 - 61; http://dx.doi.org/10.1002/ibd.20330; PMID: 18050295
- Swidsinski A, Loening-Baucke V, Verstraelen H, Osowska S, Doerffel Y. Biostructure of fecal microbiota in healthy subjects and patients with chronic idiopathic diarrhea. Gastroenterology 2008; 135:568 - 79; http://dx.doi.org/10.1053/j.gastro.2008.04.017; PMID: 18570896
- Duncan SH, Louis P, Thomson JM, Flint HJ. The role of pH in determining the species composition of the human colonic microbiota. Environ Microbiol 2009; 11:2112 - 22; http://dx.doi.org/10.1111/j.1462-2920.2009.01931.x; PMID: 19397676
- Castillo M, Skene G, Roca M, Anguita M, Badiola I, Duncan SH, Flint HJ, Martín-Orúe SM. Application of 16S rRNA gene-targetted fluorescence in situ hybridization and restriction fragment length polymorphism to study porcine microbiota along the gastrointestinal tract in response to different sources of dietary fibre. FEMS Microbiol Ecol 2007; 59:138 - 46; http://dx.doi.org/10.1111/j.1574-6941.2006.00204.x; PMID: 17004993
- Bian G, Xie F, Su Y, Zhu W. [16S rRNA gene-based molecular methods to monitor clostridium cluster IV community in the colon of piglets]. Wei sheng wu xue bao =. Acta Microbiol Sin 2010; 50:1373 - 9
- Haenen D, Zhang J, Souza da Silva C, Bosch G, van der Meer IM, van Arkel J, van den Borne JJ, Pérez Gutiérrez O, Smidt H, Kemp B, et al. A diet high in resistant starch modulates microbiota composition, SCFA concentrations, and gene expression in pig intestine. J Nutr 2013; 143:274 - 83; http://dx.doi.org/10.3945/jn.112.169672; PMID: 23325922
- Oikonomou G, Teixeira AG, Foditsch C, Bicalho ML, Machado VS, Bicalho RC, Associations of Faecalibacterium Species with Health and Growth. Fecal microbial diversity in pre-weaned dairy calves as described by pyrosequencing of metagenomic 16S rDNA. Associations of Faecalibacterium species with health and growth. PLoS One 2013; 8:e63157; http://dx.doi.org/10.1371/journal.pone.0063157; PMID: 23646192
- Gong J, Forster RJ, Yu H, Chambers JR, Sabour PM, Wheatcroft R, Chen S. Diversity and phylogenetic analysis of bacteria in the mucosa of chicken ceca and comparison with bacteria in the cecal lumen. FEMS Microbiol Lett 2002; 208:1 - 7; http://dx.doi.org/10.1111/j.1574-6968.2002.tb11051.x; PMID: 11934485
- Bjerrum L, Engberg RM, Leser TD, Jensen BB, Finster K, Pedersen K. Microbial community composition of the ileum and cecum of broiler chickens as revealed by molecular and culture-based techniques. Poult Sci 2006; 85:1151 - 64; PMID: 16830854
- Gong J, Si W, Forster RJ, Huang R, Yu H, Yin Y, Yang C, Han Y. 16S rRNA gene-based analysis of mucosa-associated bacterial community and phylogeny in the chicken gastrointestinal tracts: from crops to ceca. FEMS Microbiol Ecol 2007; 59:147 - 57; http://dx.doi.org/10.1111/j.1574-6941.2006.00193.x; PMID: 17233749
- Scupham AJ. Succession in the intestinal microbiota of preadolescent turkeys. FEMS Microbiol Ecol 2007; 60:136 - 47; http://dx.doi.org/10.1111/j.1574-6941.2006.00245.x; PMID: 17284250
- Lund M, Bjerrum L, Pedersen K. Quantification of Faecalibacterium prausnitzii- and Subdoligranulum variabile-like bacteria in the cecum of chickens by real-time PCR. Poult Sci 2010; 89:1217 - 24; http://dx.doi.org/10.3382/ps.2010-00653; PMID: 20460669
- Gérard P, Brézillon C, Quéré F, Salmon A, Rabot S. Characterization of cecal microbiota and response to an orally administered lactobacillus probiotic strain in the broiler chicken. J Mol Microbiol Biotechnol 2008; 14:115 - 22; http://dx.doi.org/10.1159/000106090; PMID: 17957118
- Meimandipour A, Shuhaimi M, Soleimani AF, Azhar K, Hair-Bejo M, Kabeir BM, Javanmard A, Muhammad Anas O, Yazid AM. Selected microbial groups and short-chain fatty acids profile in a simulated chicken cecum supplemented with two strains of Lactobacillus. Poult Sci 2010; 89:470 - 6; http://dx.doi.org/10.3382/ps.2009-00495; PMID: 20181862
- Nava GM, Stappenbeck TS. Diversity of the autochthonous colonic microbiota. Gut Microbes 2011; 2:99 - 104; http://dx.doi.org/10.4161/gmic.2.2.15416; PMID: 21694499
- Foglesong MA, Cruden DL, Markovetz AJ. Pleomorphism of fusobacteria isolated from the cockroach hindgut. J Bacteriol 1984; 158:474 - 80; PMID: 6144663
- Li M, Wang B, Zhang M, Rantalainen M, Wang S, Zhou H, Zhang Y, Shen J, Pang X, Zhang M, et al. Symbiotic gut microbes modulate human metabolic phenotypes. Proc Natl Acad Sci U S A 2008; 105:2117 - 22; http://dx.doi.org/10.1073/pnas.0712038105; PMID: 18252821
- Macfarlane GT, Macfarlane S. Fermentation in the human large intestine: its physiologic consequences and the potential contribution of prebiotics. J Clin Gastroenterol 2011; 45:Suppl S120 - 7; http://dx.doi.org/10.1097/MCG.0b013e31822fecfe; PMID: 21992950
- Gałecka M, Szachta P, Bartnicka A, Łykowska-Szuber L, Eder P, Schwiertz A. Faecalibacterium prausnitzii and Crohn’s disease - is there any connection?. Pol J Microbiol 2013; 62:91 - 5; PMID: 23829084
- Barcenilla A, Pryde SE, Martin JC, Duncan SH, Stewart CS, Henderson C, Flint HJ. Phylogenetic relationships of butyrate-producing bacteria from the human gut. Appl Environ Microbiol 2000; 66:1654 - 61; http://dx.doi.org/10.1128/AEM.66.4.1654-1661.2000; PMID: 10742256
- Duncan SH, Louis P, Flint HJ. Lactate-utilizing bacteria, isolated from human feces, that produce butyrate as a major fermentation product. Appl Environ Microbiol 2004; 70:5810 - 7; http://dx.doi.org/10.1128/AEM.70.10.5810-5817.2004; PMID: 15466518
- Duncan SH, Holtrop G, Lobley GE, Calder AG, Stewart CS, Flint HJ. Contribution of acetate to butyrate formation by human faecal bacteria. Br J Nutr 2004; 91:915 - 23; http://dx.doi.org/10.1079/BJN20041150; PMID: 15182395
- Duncan SH, Barcenilla A, Stewart CS, Pryde SE, Flint HJ. Acetate utilization and butyryl coenzyme A (CoA):acetate-CoA transferase in butyrate-producing bacteria from the human large intestine. Appl Environ Microbiol 2002; 68:5186 - 90; http://dx.doi.org/10.1128/AEM.68.10.5186-5190.2002; PMID: 12324374
- Sarbini SR, Kolida S, Gibson GR, Rastall RA. In vitro fermentation of commercial α-gluco-oligosaccharide by faecal microbiota from lean and obese human subjects. Br J Nutr 2013; 109:1980 - 9; http://dx.doi.org/10.1017/S0007114512004205; PMID: 23116939
- Sarbini SR, Kolida S, Naeye T, Einerhand A, Brison Y, Remaud-Simeon M, Monsan P, Gibson GR, Rastall RA. In vitro fermentation of linear and alpha-1,2-branched dextrans by the human fecal microbiota. Appl Environ Microbiol 2011; 77:5307 - 15; http://dx.doi.org/10.1128/AEM.02568-10; PMID: 21666027
- Dabek M, McCrae SI, Stevens VJ, Duncan SH, Louis P. Distribution of beta-glucosidase and beta-glucuronidase activity and of beta-glucuronidase gene gus in human colonic bacteria. FEMS Microbiol Ecol 2008; 66:487 - 95; http://dx.doi.org/10.1111/j.1574-6941.2008.00520.x; PMID: 18537837
- Benjdia A, Martens EC, Gordon JI, Berteau O. Sulfatases and a radical S-adenosyl-L-methionine (AdoMet) enzyme are key for mucosal foraging and fitness of the prominent human gut symbiont, Bacteroides thetaiotaomicron. J Biol Chem 2011; 286:25973 - 82; http://dx.doi.org/10.1074/jbc.M111.228841; PMID: 21507958
- Sonnenburg JL, Xu J, Leip DD, Chen CH, Westover BP, Weatherford J, Buhler JD, Gordon JI. Glycan foraging in vivo by an intestine-adapted bacterial symbiont. Science 2005; 307:1955 - 9; http://dx.doi.org/10.1126/science.1109051; PMID: 15790854
- Maccaferri S, Vitali B, Klinder A, Kolida S, Ndagijimana M, Laghi L, Calanni F, Brigidi P, Gibson GR, Costabile A. Rifaximin modulates the colonic microbiota of patients with Crohn’s disease: an in vitro approach using a continuous culture colonic model system. J Antimicrob Chemother 2010; 65:2556 - 65; http://dx.doi.org/10.1093/jac/dkq345; PMID: 20852272
- Dörffel Y, Swidsinski A, Loening-Baucke V, Wiedenmann B, Pavel M. Common biostructure of the colonic microbiota in neuroendocrine tumors and Crohn’s disease and the effect of therapy. Inflamm Bowel Dis 2012; 18:1663 - 71; http://dx.doi.org/10.1002/ibd.21923; PMID: 22113988
- Bartosch S, Fite A, Macfarlane GT, McMurdo ME. Characterization of bacterial communities in feces from healthy elderly volunteers and hospitalized elderly patients by using real-time PCR and effects of antibiotic treatment on the fecal microbiota. Appl Environ Microbiol 2004; 70:3575 - 81; http://dx.doi.org/10.1128/AEM.70.6.3575-3581.2004; PMID: 15184159
- Clavel T, Fallani M, Lepage P, Levenez F, Mathey J, Rochet V, Sérézat M, Sutren M, Henderson G, Bennetau-Pelissero C, et al. Isoflavones and functional foods alter the dominant intestinal microbiota in postmenopausal women. J Nutr 2005; 135:2786 - 92; PMID: 16317121
- Thapa D, Losa R, Zweifel B, Wallace RJ. Sensitivity of pathogenic and commensal bacteria from the human colon to essential oils. Microbiology 2012; 158:2870 - 7; http://dx.doi.org/10.1099/mic.0.061127-0; PMID: 22878397
- Zwielehner J, Lassl C, Hippe B, Pointner A, Switzeny OJ, Remely M, Kitzweger E, Ruckser R, Haslberger AG. Changes in human fecal microbiota due to chemotherapy analyzed by TaqMan-PCR, 454 sequencing and PCR-DGGE fingerprinting. PLoS One 2011; 6:e28654; http://dx.doi.org/10.1371/journal.pone.0028654; PMID: 22194876
- Benus RF, van der Werf TS, Welling GW, Judd PA, Taylor MA, Harmsen HJ, Whelan K. Association between Faecalibacterium prausnitzii and dietary fibre in colonic fermentation in healthy human subjects. Br J Nutr 2010; 104:693 - 700; http://dx.doi.org/10.1017/S0007114510001030; PMID: 20346190
- Shen Q, Zhao L, Tuohy KM. High-level dietary fibre up-regulates colonic fermentation and relative abundance of saccharolytic bacteria within the human faecal microbiota in vitro. Eur J Nutr 2012; 51:693 - 705; http://dx.doi.org/10.1007/s00394-011-0248-6; PMID: 21952691
- Fernando WM, Hill JE, Zello GA, Tyler RT, Dahl WJ, Van Kessel AG. Diets supplemented with chickpea or its main oligosaccharide component raffinose modify faecal microbial composition in healthy adults. Benef Microbes 2010; 1:197 - 207; http://dx.doi.org/10.3920/BM2009.0027; PMID: 21831757
- Hooda S, Boler BM, Serao MC, Brulc JM, Staeger MA, Boileau TW, Dowd SE, Fahey GC Jr., Swanson KS. 454 pyrosequencing reveals a shift in fecal microbiota of healthy adult men consuming polydextrose or soluble corn fiber. J Nutr 2012; 142:1259 - 65; http://dx.doi.org/10.3945/jn.112.158766; PMID: 22649263
- Bassaganya-Riera J, Hontecillas R. Dietary conjugated linoleic acid and n-3 polyunsaturated fatty acids in inflammatory bowel disease. Curr Opin Clin Nutr Metab Care 2010; 13:569 - 73; http://dx.doi.org/10.1097/MCO.0b013e32833b648e; PMID: 20508519
- Jia W, Whitehead RN, Griffiths L, Dawson C, Waring RH, Ramsden DB, Hunter JO, Cole JA. Is the abundance of Faecalibacterium prausnitzii relevant to Crohn’s disease?. FEMS Microbiol Lett 2010; 310:138 - 44; http://dx.doi.org/10.1111/j.1574-6968.2010.02057.x; PMID: 20695899
- Dewulf EM, Cani PD, Claus SP, Fuentes S, Puylaert PG, Neyrinck AM, Bindels LB, de Vos WM, Gibson GR, Thissen JP, et al. Insight into the prebiotic concept: lessons from an exploratory, double blind intervention study with inulin-type fructans in obese women. Gut 2013; 62:1112 - 21; http://dx.doi.org/10.1136/gutjnl-2012-303304; PMID: 23135760
- Louis P, Flint HJ. Diversity, metabolism and microbial ecology of butyrate-producing bacteria from the human large intestine. FEMS Microbiol Lett 2009; 294:1 - 8; http://dx.doi.org/10.1111/j.1574-6968.2009.01514.x; PMID: 19222573
- Ramirez-Farias C, Slezak K, Fuller Z, Duncan A, Holtrop G, Louis P. Effect of inulin on the human gut microbiota: stimulation of Bifidobacterium adolescentis and Faecalibacterium prausnitzii. Br J Nutr 2009; 101:541 - 50; http://dx.doi.org/10.1017/S0007114508019880; PMID: 18590586
- Salazar N, Gueimonde M, Hernández-Barranco AM, Ruas-Madiedo P, de los Reyes-Gavilán CG. Exopolysaccharides produced by intestinal Bifidobacterium strains act as fermentable substrates for human intestinal bacteria. Appl Environ Microbiol 2008; 74:4737 - 45; http://dx.doi.org/10.1128/AEM.00325-08; PMID: 18539803
- Odamaki T, Xiao JZ, Iwabuchi N, Sakamoto M, Takahashi N, Kondo S, Miyaji K, Iwatsuki K, Togashi H, Enomoto T, et al. Influence of Bifidobacterium longum BB536 intake on faecal microbiota in individuals with Japanese cedar pollinosis during the pollen season. J Med Microbiol 2007; 56:1301 - 8; http://dx.doi.org/10.1099/jmm.0.47306-0; PMID: 17893165
- Garrido D, Suau A, Pochart P, Cruchet S, Gotteland M. Modulation of the fecal microbiota by the intake of a Lactobacillus johnsonii La1-containing product in human volunteers. FEMS Microbiol Lett 2005; 248:249 - 56; http://dx.doi.org/10.1016/j.femsle.2005.05.045; PMID: 15970400