Abstract
Here, we report on GEO-D03, a DNA vaccine that co-expresses non-infectious HIV-1 virus-like particles (VLPs) and the human cytokine, granulocyte-macrophage colony-stimulating factor (GM-CSF). The virus-like particles display the native gp160 form of the HIV-1 Envelope glycoprotein (Env) and are designed to elicit antibody against the natural form of Env on virus and virus-infected cells. The DNA-expressed HIV Gag, Pol and Env proteins also have the potential to elicit virus-specific CD4 and CD8 T cells. The purpose of the co-expressed GM-CSF is to target a cytokine that recruits, expands and differentiates macrophages and dendritic cells to the site of VLP expression. The GEO-D03 DNA vaccine is currently entered into human trials as a prime for a recombinant modified vaccinia Ankara (MVA) boost. In preclinical studies in macaques using an SIV prototype vaccine, this vaccination regimen elicited both anti-viral T cells and antibody, and provided 70% protection against acquisition during 12 weekly rectal exposures with a heterologous SIV. Higher avidity of the Env-specific Ab for the native form of the Env in the challenge virus correlated with lower likelihood of SIV infection.
GEO-D03, a DNA vaccine co-expressing HIV-1 virus-like particles and granulocyte-macrophage colony-stimulating factor (GM-CSF), has been designed to prime immune responses against HIV. Precedents for the use of VLP in recombinant vaccines can be found in the two licensed genetically engineered recombinant viral vaccines: those for hepatitis B and human papilloma viruses, consisting of virus-like particles.Citation1,Citation2 For both of these vaccines, VLPs are produced using recombinant DNA technology. Precedence for the ability of GM-CSF to enhance vaccine responses is found in Provenge, the first licensed cancer vaccine.Citation3 In Provenge, a fusion of GM-CSF with prostatic acid phosphatase is used to load autologous dendritic cells ex vivo. The treated dendritic cells are then infused to elicit anti-tumor immune responses. In our macaque trials testing the protective potential of prototype HIV vaccines, co-expression of GM-CSF in the DNA prime for a recombinant MVA boost has proven to be highly beneficial for preventing infection by repeated heterologous rectal exposures, increasing prevention of infection from 25% to 71%.Citation4
GEO-D03 was constructed by expressing GM-CSF in the position of nef in the pGA2/JS7 DNA vaccine that expresses VLP. pGA2/JS7 has undergone Phase 1 testing as a prime for a modified vaccinia Ankara (MVA) boostCitation5 and is currently undergoing Phase 2a testing in humans. The pGA2/JS7 DNA vaccine is a 9.5 kb plasmid DNA composed of a 2.9 kb expression vector termed pGA2 and a 6.6 kb vaccine insert termed JS7. The JS7 vaccine insert expresses the HIV-1 proteins Gag, protease (PR), reverse transcriptase (RT), gp160 Env, Tat, Vpu, and Rev from a single transcript that undergoes subgenomic splicing in mammalian cells.Citation6 The subgenomic splicing is rev-dependent and uses the rev responsive element and splicing signals present in the wild type HIV-1 genome. The VLP have been rendered noninfectious by deletions as well as point mutations.Citation6,Citation7 The deleted regions include both long-terminal repeats, a portion of the 5′ sequences encoding encapsidation of viral RNA, and coding sequences for integrase, Vif, Vpr, and Nef. Point mutations reduce packaging of viral RNA and inactivate the viral PR, RT, strand transfer, and RNase H activities encoded in Pol. Expression of GM-CSF in JS7 in the position of Nef was achieved using a synthetic sequence and standard recombinant DNA technology.
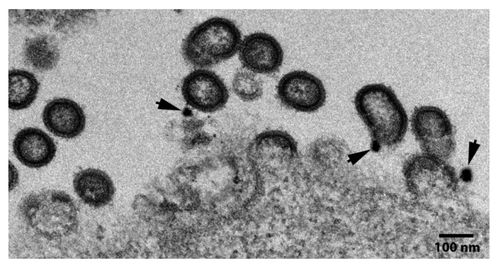
Electron micrographs of 293T cells transiently transfected with GEO-D03 DNA revealed the anticipated expression of immature virus-like particles (). The VLPs were observed budding from cells and adjacent to cells. Immunogold staining for the presence of Env using a mixture of 4 rabbit monoclonals to gp120 revealed the presence of Env on the surface of the particles (see arrow heads, ).
Figure 1. Electron micrograph showing GEO-D03 production of Env-displaying VLP. Arrows show immunogold staining of Env. GEO-D03-transfected HEK293T cells were fixed at ~48 h post transfection with 4% paraformaldehyde in 0.1 M phosphate buffer (pH 7.4) and treated with 0.1% sodium borohydride followed by permeabilization with 0.05% Triton X-100 in phosphate buffer. Potential non-specific binding was blocked with 5% normal goat serum, 5% bovine serum albumin (BSA), and 0.1% gelatin. Cells were then incubated with a mixture of four rabbit anti-Env monoclonal antibodies (# 13, 53, 56, and 76). After washing, cells were incubated with secondary goat anti-rabbit antibody conjugated to Ultrasmall gold particles (Aurion, the Netherlands) for 12 h at 4°C. Silver enhancement was conducted and cells were fixed with 0.5% osmium tetroxide for 30 min followed by dehydration and embedding in Epoxy resin according to the conventional method. Ultrathin sections were cut at 70 nm, stained with 4% aquous uranyl acetate and 2% lead citrate, and examined on a JEOL JEM-1400 transmission electron microscope (JEOL USA, Peabody, MA) equipped with a Gatan US1000.894 Ultrascan CCD camera (Gatan, Inc., Pleasanton CA).
Like HIV-1 proviral DNA, the GEO-D03 DNA vaccine expresses multiple proteins from a single transcript through alternative splicing and alternate reading frames (). Full-length RNA is spliced into two subgenomic classes of mRNAs: 4.0-kb, and 1.8-kb.Citation8 Because of the presence of multiple closely spaced splice donor and acceptor sites, subgenomic mRNAs are grouped into size classes. Based on the deletions and putative splice junctions in the GEO-D03 sequence, we predicted full length mRNA at 7.3 kb and subgenomic mRNAs at ~3.4 kb, and ~1.4 kb. If processed correctly, the full-length 7.3 kb mRNA expresses Gag-Pol, the 3.4 kb subclass mRNA expresses Vpu and Env, and the 1.8 kb subclass mRNA expresses Tat, Rev, and GM-CSF. However, all of the subclasses of mRNA include GM-CSF sequences at the 3′ end of their mRNA.
Figure 2. northern blot analyses of GEO-D03-expressed mRNAS. (A) Schematic showing reading frames for proteins expressed by GEO-D03 DNA (open rectangles) and size classes of spliced mRNAs (filled rectangles). The asterisks indicate multiple, closely spaced spliced sites. (B) Northern blot probed for GM-CSF sequences. The sequences in each size class of mRNA and the proteins expressed by each size class are shown to the right of the blot. GEO-D03 DNA and pGA2/JS7 were transfected into HEK293T cells in six-well plates at 1 µg DNA per well using the LipofectAmine 2000 transfection reagent (Life Technologies). After a two-day incubation, cells were trypsinized, and RNA was purified from the cells with the RNAqueous kit (Life Technologies) according to the kit protocol. Electrophoresis and transfer were performed using the NorthernMax kit (Life Technologies) according to the kit protocol. The biotinylated Gag probe was hybridized to the membrane and the blot processed according to standard procedures and scanned on an Odyssey infrared scanner (Li-Cor). Once scanned, the blot was stripped with 0.5% SDS and re-probed with the GM-CSF probe.
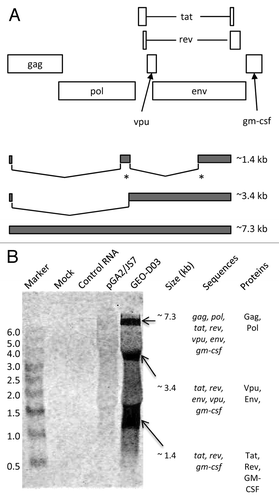
Northern blot analyses revealed the correct transcription and splicing of the GEO-D03 message to create properly-processed mRNAs (). Splicing patterns were determined in total RNA purified from HEK293T cells transfected with GEO-D03 or JS7 DNA. Probes were generated through PCR amplification of fragments of gag and gm-csf . On the Gag blot, the gag sequence was detected at approximately 7.3 kb in cells transfected with GEO-D03 and at approximately 6.9 kb in cells transfected with pGA2/JS7 (data not shown). This result is consistent with the predicted splicing patterns (), in which the gag sequence is present in the full-length mRNA but spliced out of the 3.4- and 1.8-kb classes. On the GM-CSF blot, the GM-CSF sequence was detected in all three classes of mRNA from cells transfected with GEO-D03 consistent with the predicted splicing pattern (), in which the gm-csf sequence is present in all classes of mRNA. The gm-csf sequence was not detected in RNA from cells transfected with JS7 DNA ().
Western blots demonstrated that GEO-D03, like its precursor JS7, expresses full-length Gag and Env proteins (). Furthermore, the blots show that GEO-D03, but not JS7, expresses full-length GM-CSF, which was glycosylated as expected (). Western blot analyses were performed on lysates and supernatants of cells transfected with GEO-D03 DNA, JS7 DNA and mock-transfected cells. In the Gag blot, the full-length pr55 protein is clearly present in lysates and supernatants from cells transfected with pGA2/JS7 and GEO-D03 DNA vaccines. Similarly, the Env blot demonstrated expression of the full-length gp160 protein as well as cleavage of gp160 to the gp120 form. As expected, the gp120 band is more prominent in supernatants than in lysates, indicating greater processing of secreted protein. Migration patterns are similar from both plasmids; therefore, addition of the GM-CSF sequence to the JS7 plasmid did not affect the ability to express full-length Gag and Env proteins.
Figure 3. western blot analyses of proteins expressed by GEO-D03. (A) Expressed Gag proteins, (B) expressed Env proteins, and (C). Expressed GM-CSF. HEK293T cells were transiently transfected with GEO-D03 or pGA2/JS7 DNA. Lysates and supernatants harvested at 48 h were separated by SDS-PAGE according to standard methods. 4–15% gradient SDS-PAGE gels (Bio-Rad) were used for Gag and Env detection; and a 12.5% SDS-PAGE gel (Bio-Rad), for GM-CSF detection. Recombinant, E. coli-derived GM-CSF protein was loaded as a control on the GM-CSF gel. Gag was detected with the H12–5C mouse monoclonal antibody, and Env with the T24 mouse monoclonal antibody; both antibodies were obtained from the NIH AIDS Research and Reference Reagent Program. GM-CSF was detected with the B6–2-hGMCSF mouse monoclonal antibody (Santa Cruz Biotechnology). For detection of bound antibody, all membranes were probed with anti-mouse-AlexaFluor 680 (Life Technologies) and then scanned on an Odyssey infrared scanner (Li-Cor). D03, GEO-D03; Sup, supernatant; Std, standard; Lys, lysate.
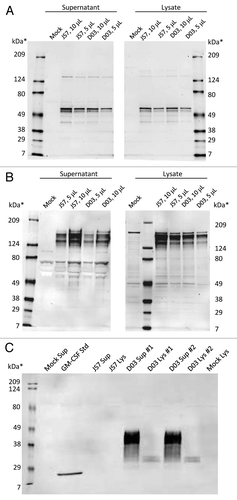
In the GEO-D03 lanes of the GM-CSF blot, expression bands are present at approximately 30 kDa in lysates and at approximately 36 kDa in supernatants, whereas the E. coli-derived GM-CSF has an approximate molecular weight of 17 kDa. The molecular weight of the E. coli-derived GM-CSF corresponds well with the predicted mass of 16.2 kDa based on its primary sequence. The much higher molecular weights of the proteins from transfected cells are due to extensive glycosylation of GM-CSF in mammalian cells. GM-CSF expression was not seen in lanes from cells transfected with pGA2/JS7.
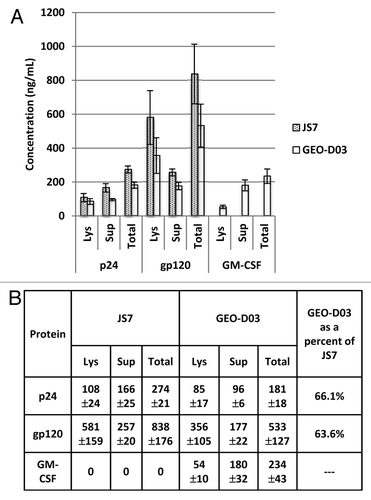
In addition to the western blot analysis to confirm expression of full-length Gag, Env, and GM-CSF proteins, we performed ELISA analyses to quantify expression of these proteins (). The ELISA data demonstrate that GEO-D03 expresses Gag and Env proteins in quantities that are 60–70% of the quantities expressed by pGA2/JS7. We hypothesize that the lower expression of Gag and Env from GEO-D03 is due to the subgenomic splicing of its mRNA including the formation of the 1.4 kb GM-CSF mRNA. The data also indicate that GM-CSF is expressed at approximately 200 nanograms per 106 cells, per day, a level that has proven effective as an adjuvant for protection in our studies in macaques.Citation4 As expected, the Gag and Env proteins were detected in lysates and supernatants from cells transfected with pGA2/JS7 and GEO-D03, whereas GM-CSF was detected only in lysates and supernatants from cells transfected with GEO-D03. While Env is present in greater quantities in lysates than in supernatants, Gag and GM-CSF were present at higher levels in supernatants than in lysates. The ratio of secreted-to-intracellular protein is especially high for GM-CSF. Exact molar quantities of the proteins cannot be calculated accurately due to the unknown affinity of the kit antibodies for the vaccine-expressed proteins; however, general conclusions may be drawn. The calculated quantity of gp120 Env protein was approximately four to five times as high as the calculated quantity of p24 Gag protein. Because of the higher molecular weight of Env relative to Gag, the molar ratio of Env to Gag is likely closer to one.
Figure 4. Quantification of Gag, Env and GM-CSF expression by ELISA. (A) Expression of p24 Gag, gp120 Env and GM-CSF in lysates and supernatants. (B) Summary of Expression data. HEK293T cells were transiently transfected with GEO-D03, pGA2/JS7, or mock transfected. After a two-day incubation, Gag, Env, and GM-CSF proteins in lysates and supernatants were quantified using the p24 Antigen Capture Assay Kit (ABL), the HIV-1 gp120 Clade B specific Antigen Capture Assay Kit (ABL), and the ELISA for Human GM-CSF Kit (Mabtech), respectively. Values are ng of protein produced by approximately 1 × 106 transfected cells expressed as mean ± standard deviation. Lys, lysate; Sup, supernatant.
GEO-D03 vaccine DNA, used as a prime for an MVA boost, was first inoculated into humans on May 18th 2012, in a dose-escalation study through the HIV Vaccine Trials Network. The doses of DNA are the same as those used in the dose escalation of its parent plasmid pGA2/JS2.Citation5 The 1/10th dose is 0.3 mg, and full dose is 3 mg. All inoculations are intramuscular with needle and syringe.
In the non-human primate model, priming with an SIV prototype of the GEO-D03 DNA and boosting with MVA has proven particularly effective at eliciting protective immunity.Citation4 With the GM-CSF co-expressing DNA prime, the vaccine achieved a 90% reduction in risk of infection with 70% of the macaques protected against 12 weekly rectal challenges. In the absence of the co-expressed GM-CSF, the vaccine achieved a 61% reduction in the relative risk of infection with 25% of the macaques protected against infection during the 12 weekly exposures.Citation4 Co-expression of GM-CSF in the DNA prime enhanced neutralizing antibody titers, antibody-dependent cellular cytotoxicity and the avidity of Env-specific antibody.Citation4 Use of GM-CSF also enhanced the frequency of detection of viral-specific IgA in rectal secretions.Citation4 Of these parameters, the avidity of Env-specific antibody for the Env of the challenge virus, an overall measure of the quality of the Ab response for the Env of the challenge virus, correlated with prevention of infection (r = 0.9, p = < 0.0001). Neutralizing antibody, antibody-dependent cellular cytotoxicity and viral-specific rectal IgA did not correlate with protection. However, the individual Envs used in these latter tests may not have accurately mimicked the Envs undergoing transmission
We believe that GEO-D03 effectively primes protective immunity due largely to the presence of two features:Citation6,Citation7 the expression of VLP displaying full length Env andCitation9,Citation10 the co-expression of GM-CSF. Virus-like particles have been found effective as vaccines, presumably because VLP mimic viruses in their natural form. We also consider display of the native trimeric gp160 form of Env on the surface of the VLPs to be important. We have found that priming and boosting with MVA that expresses VLPs displaying a truncated trimeric gp150 Env elicits higher titer and higher avidity Ab than priming with DNA that expresses gp160 Env and boosting with MVA that expresses gp150 Env.Citation11 Disappointingly, the avidity of this Ab did not correlate with prevention of infection, a phenomenon that we believe reflects the trimeric gp150 Env failing to elicit Ab against the native gp160 structure of Env.Citation11 The C-terminal complete gp160 Env is toxic to cellsCitation12 and difficult to express in live recombinant vaccine vectors.Citation13 However it is possible to express gp160 Env using recombinant DNA. Thus, the DNA prime affords a unique opportunity to express VLPs that display the complete gp160 form of Env.
We also consider the ability of GEO-D03 to co-express GM-CSF at the site where VLPs are being presented to the immune system important. GM-CSF stimulates the production and differentiation of monocytes into dendritic cells and macrophages. In turn, dendritic cells and macrophages initiate adaptive immune responses (for review, see.Citation10 GM-CSF is licensed for human use for acceleration of white blood cell recovery following bone marrow transplantation, for treatment of fungal infections, for replacement of white blood cells following chemotherapy and, most recently, for use in a fusion protein as an adjuvant for a dendritic cell therapy for prostate cancer. GM-CSF has been broadly used in preclinical vaccine studies. It was the first reported “genetic adjuvant” for DNA vaccines.Citation14 GM-CSF stood out in early preclinical screens of retroviral expressed cytokines as an adjuvant for cancer cell vaccines,Citation15 and can serve as an effective adjuvant for immunizations with peptides.Citation9
There are also caveats for the use of GM-CSF as a vaccine adjuvant. The level of GM-CSF expression is important. If GM-CSF is present at excessive levels, it expands myeloid suppressor cells that dampen adaptive immune responses.Citation16-Citation18 The context in which GM-CSF is expressed is also important for eliciting protective immunity for immunodeficiency virus infections. GM-CSF expressed in recombinant vesicular stomatitis SIV vaccine and recombinant MVA vaccine simian immunodeficiency virus vaccines have not enhanced protection in preclinical macaque studies (ref. Citation19 and Amara and Robinson, unpublished observations).
In summary, we are pleased to report the construction and expression testing of GEO-D03, a DNA prime that co-expresses GM-CSF and virus-like particles. The goal of our recently initiated human trials is to determine whether priming with GEO-D03 DNA and boosting with MVA will achieve the excellent protection in humans that was achieved by its SIV-prototype in macaques.
Acknowledgments
This research was supported an Integrated Preclinical/Clinical AIDS Vaccine Development program project from the US Public Health Service, U19AI074073. We would like to thank Susan Reuland for expert administrative assistance.
Disclosure of Potential Conflicts of Interest
Michael Hellerstein, Yongxian Xu and Harriet Robinson are employees of GeoVax, Inc., a compmany that is developing GEO-D03 as a prime for a commercial DNA/MVA HIV vaccine.
References
- Mast E, Mahoney F, Kane M, Margolis H. Hepatitis b vaccine. Vaccines 2004:Chapter 16, Philadelphia, PA, W.B. Saunders Co.
- Schiller JT, Lowy DR. Human papillomavirus vaccine for cervical cancer prevention. Vaccines 2004; Chapter 45, Philadelphia, PA, W.B. Saunders Co.
- Small EJ, Fratesi P, Reese DM, Strang G, Laus R, Peshwa MV, et al. Immunotherapy of hormone-refractory prostate cancer with antigen-loaded dendritic cells. J Clin Oncol 2000; 18:3894 - 903; PMID: 11099318
- Lai L, Kwa S, Kozlowski PA, Montefiori DC, Ferrari G, Johnson WE, et al. Prevention of infection by a granulocyte-macrophage colony-stimulating factor co-expressing DNA/modified vaccinia Ankara simian immunodeficiency virus vaccine. J Infect Dis 2011; 204:164 - 73; http://dx.doi.org/10.1093/infdis/jir199; PMID: 21628671
- Goepfert PA, Elizaga ML, Sato A, Qin L, Cardinali M, Hay CM, et al, National Institute of Allergy and Infectious Diseases HIV Vaccine Trials Network. Phase 1 safety and immunogenicity testing of DNA and recombinant modified vaccinia Ankara vaccines expressing HIV-1 virus-like particles. J Infect Dis 2011; 203:610 - 9; http://dx.doi.org/10.1093/infdis/jiq105; PMID: 21282192
- Smith JM, Amara RR, Campbell D, Xu Y, Patel M, Sharma S, et al. DNA/MVA vaccine for HIV type 1: effects of codon-optimization and the expression of aggregates or virus-like particles on the immunogenicity of the DNA prime. AIDS Res Hum Retroviruses 2004; a 20:1335 - 47; http://dx.doi.org/10.1089/aid.2004.20.1335; PMID: 15650426
- Smith JM, Amara RR, McClure HM, Patel M, Sharma S, Yi H, et al. Multiprotein HIV-1 clade b DNA/MVA vaccine: construction, safety and immunogenicity. AIDS Res Hum Retro 2004; b 20:654 - 65; http://dx.doi.org/10.1089/0889222041217419
- Purcell DF, Martin MA. Alternative splicing of human immunodeficiency virus type 1 mRNA modulates viral protein expression, replication, and infectivity. J Virol 1993; 67:6365 - 78; PMID: 8411338
- Berzofsky JA, Ahlers JD, Belyakov IM. Strategies for designing and optimizing new generation vaccines. Nat Rev Immunol 2001; 1:209 - 19; http://dx.doi.org/10.1038/35105075; PMID: 11905830
- Borrello I, Pardoll D. GM-CSF-based cellular vaccines: a review of the clinical experience. Cytokine Growth Factor Rev 2002; 13:185 - 93; http://dx.doi.org/10.1016/S1359-6101(01)00034-X; PMID: 11900993
- Lai L, Kwa SF, Kozlowski PA, Montefiori DC, Nolen TL, Hudgens MG, et al. SIVmac239 MVA vaccine with and without a DNA prime, similar prevention of infection by a repeated dose SIVsmE660 challenge despite different immune responses. Vaccine 2012; 30:1737 - 45; http://dx.doi.org/10.1016/j.vaccine.2011.12.026; PMID: 22178526
- Miller MA, Mietzner TA, Cloyd MW, Robey WG, Montelaro RC. Identification of a calmodulin-binding and inhibitory peptide domain in the HIV-1 transmembrane glycoprotein. AIDS Res Hum Retroviruses 1993; 9:1057 - 66; http://dx.doi.org/10.1089/aid.1993.9.1057; PMID: 8312049
- Wyatt LS, Belyakov IM, Earl PL, Berzofsky JA, Moss B. Enhanced cell surface expression, immunogenicity and genetic stability resulting from a spontaneous truncation of HIV Env expressed by a recombinant MVA. Virology 2008; 372:260 - 72; http://dx.doi.org/10.1016/j.virol.2007.10.033; PMID: 18048074
- Xiang Z, Ertl HC. Manipulation of the immune response to a plasmid-encoded viral antigen by coinoculation with plasmids expressing cytokines. Immunity 1995; 2:129 - 35; http://dx.doi.org/10.1016/S1074-7613(95)80001-8; PMID: 7895169
- Dranoff G, Jaffee E, Lazenby A, Golumbek P, Levitsky H, Brose K, et al. Vaccination with irradiated tumor cells engineered to secrete murine granulocyte-macrophage colony-stimulating factor stimulates potent, specific, and long-lasting anti-tumor immunity. Proc Natl Acad Sci U S A 1993; 90:3539 - 43; http://dx.doi.org/10.1073/pnas.90.8.3539; PMID: 8097319
- Serafini P, Carbley R, Noonan KA, Tan G, Bronte V, Borrello I. High-dose granulocyte-macrophage colony-stimulating factor-producing vaccines impair the immune response through the recruitment of myeloid suppressor cells. Cancer Res 2004; 64:6337 - 43; http://dx.doi.org/10.1158/0008-5472.CAN-04-0757; PMID: 15342423
- Filipazzi P, Valenti R, Huber V, Pilla L, Canese P, Iero M, et al. Identification of a new subset of myeloid suppressor cells in peripheral blood of melanoma patients with modulation by a granulocyte-macrophage colony-stimulation factor-based antitumor vaccine. J Clin Oncol 2007; 25:2546 - 53; http://dx.doi.org/10.1200/JCO.2006.08.5829; PMID: 17577033
- Parmiani G, Castelli C, Pilla L, Santinami M, Colombo MP, Rivoltini L. Opposite immune functions of GM-CSF administered as vaccine adjuvant in cancer patients. Ann Oncol 2007; 18:226 - 32; http://dx.doi.org/10.1093/annonc/mdl158; PMID: 17116643
- Schell JB, Bahl K, Rose NF, Buonocore L, Hunter M, Marx PA, et al. Viral vectored granulocyte-macrophage colony stimulating factor inhibits vaccine protection in an SIV challenge model: protection correlates with neutralizing antibody. Vaccine 2012; 30:4233 - 9; http://dx.doi.org/10.1016/j.vaccine.2012.04.046; PMID: 22537983