Abstract
Nosocomial infections, also called “hospital acquired infections,” occur worldwide and affect both developed and resource-poor countries, thus having a major impact on their health care systems. Klebsiella pneumoniae, which is an opportunistic Gram-negative pathogen, is responsible for causing pneumonia, urinary tract infections and septicemia in immune compromised hosts such as neonates. Unfortunately, there is no vaccine or mAb available for prophylactic or therapeutic use against K. pneumoniae infections. For this reason, we sought for a protein-based subunit vaccine capable of combating K. pneumoniae infections, by applying our ANTIGENome technology for the identification of potential vaccine candidates, focusing on conserved protein antigens present in strains with different serotypes. We identified numerous novel immunogenic proteins using genomic surface display libraries and human serum antibodies from donors exposed to or infected by K. pneumoniae. Vaccine candidate antigens were finally selected based on animal protection in a murine lethal-sepsis model. The protective and highly conserved antigens identified in this study are promising candidates for the development of a protein-based vaccine to prevent infection by K. pneumoniae.
Introduction
K. pneumoniae is a common hospital-acquired pathogen, causing urinary tract infections, nosocomial pneumonia, intra-abdominal infections, surgical wound infections and infections of the blood. All of these infections can progress to shock and death if not treated early in an aggressive fashion. K. pneumoniae is also a potential community-acquired pathogen. It is estimated that Klebsiella spp account for 8% of endemic hospital infections and 3% of epidemic outbreaks.Citation1 A recent review of all major studies performed in developing countries conducted between 1990 and 2004 concluded that Klebsiella spp were the leading cause of serious bacterial neonatal infections in developing countries.Citation2 In a seven-year (1995–2002) surveillance study in US hospitals Klebsiella spp ranked 6th as a cause of nosocomial bloodstream infections.Citation3K. pneumoniae is among the most frequently isolated microorganism in intensive care units-acquired pneumonia,Citation22 hospital-acquired urinary tract and wound infections.
The pathogenicity of Klebsiella can be attributed to its production of a heat-stable enterotoxin. Further virulence factors of K. pneumoniae which have been identified so far include capsular polysaccharides (CPS), lipopolysaccharides, adhesins (type 1 and 3 pili, KPF-28 fimbriae, CF29K and an aggregative adhesin) and iron acquisition systems.Citation4 Klebsiella species may contain resistance plasmids (R-plasmids) which confer resistance to antibiotics such as ampicillin and carbenicillin.Citation5 To make matters worse, the R-plasmids can be transferred to other enteric bacteria of the same, but also of different species. Outbreaks of multidrug-resistant Klebsiella spp in hospitals are often caused by new ESBL (extended spectrum β-lactamase) producing strains. The prevalence of ESBL-producing strains among clinical Klebsiella isolates has steadily increased over the past several years.Citation6
Several attempts aiming to develop a vaccine against Klebsiella were reported up-to-date.Citation7-Citation10 Among the different cell constituents, two surface components are mainly being discussed as candidates for an anti-Klebsiella vaccine: LPS and CPS.Citation11,Citation12 While the utilization of LPS antigens in Klebsiella vaccines is favored by the existence of only 9 different O-types, the adverse toxic reactions present a great drawback of active immunization with LPS-containing vaccines, although they can be reduced by detoxification. CPS in contrast, has been proven to be highly immunogenic and nontoxic.Citation13 However, the serious disadvantage of a Klebsiella CPS vaccine is the great number of K-types (77 different antigens). CPS–based vaccines should be multivalent against at least the 24 major K-types, in order to cover 70% of all bacteremia isolates.Citation14 A 24-valent Klebsiella CPS vaccine was developed and subsequently shown to be safe and immunogenic,Citation15 yet no further development has been reported. To overcome the disadvantages of the above-mentioned approaches, conserved protein based vaccines against Klebsiella may provide a promising alternative. Kurupati et al.Citation16 have recently used a proteomic approach and identified a number of immunogenic K. pneumoniae antigens, included FepA, OmpA, OmpK17, OmpK36 and Colicin I receptor, which were considered as candidates for vaccine development. More recently, a study in mice showed efficacy against K. pneumoniae infection for DNA vaccines based on outer membrane proteins.Citation17
In order to identify vaccine candidates naturally recognized by the human immune system, we applied the ANTIGENome technology to K. pneumoniae for the comprehensive identification of novel conserved and protective antigens suitable for vaccine development to prevent K. pneumoniae infections.Citation18,Citation19 For immune selection, we used human serum antibodies obtained from patients recorded with disease symptoms or from individuals with previous K. pneumoniae infections. These studies led to the discovery of eight novel antigens, all of which are highly conserved among Klebsiella clinical isolates and provide significant protection in murine challenge models.
Results
Characterization and selection of human serum samples for genomic antigen screens
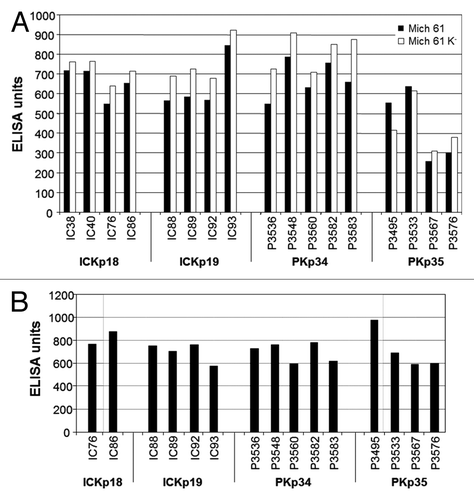
A collection of human sera from 100 patients with a confirmed medical diagnosis of K. pneumoniae infections and 89 sera from healthy individuals were characterized for antigen screening. The recorded disease symptoms and medical diagnosis of the patients included skin soft tissue infection, pneumonia, septicemia, intra-abdominal Infection and urinary tract infection. In order to select sera with a high titer of Klebsiella-specific antibodies, all serum samples were analyzed by ELISA using whole cell extracts of K. pneumoniae strains. A broad range of responses were detected (data not shown). The sera with the highest titer were selected for subsequent antigen identification and validation studies. Four different serum pools were prepared including 17 individual serum samples, with two pools from patients (PKp34 and PKp35) and two pools with sera from healthy donors (ICKp18 and ICKp19). The selected IgG pools recognized large arrays of proteins in extracts of different K. pneumoniae strains, including MGH 78578, Mich 61 and capsule negative Mich 61 mutant ( and B). Importantly, the comparison of the IgG reactivity against total bacterial lysates prepared from K. pneumoniae strain Mich 61 and its mutant showed that the antibodies did not primarily recognize capsular polysaccharide antigens ().
Figure 1. Characterization of human sera for anti-K. pneumoniae antibodies as measured by ELISA. Total anti-K. pneumoniae IgG antibody levels were measured by ELISA (A) total bacterial lysates prepared from K. pneumoniae strain Mich 61 and its respective capsule negative mutant. (B) Total bacterial lysates prepared from K. pneumoniae MGH 78578. The graph represents the ELISA units of 17 sera with the highest antibody titers against K. pneumoniae lysate. Data are expressed as ELISA units calculated from the absorbance at 405 nm at a serum dilution of 1:1,000. Human sera indication: IC, healthy individual; P, patient.
Prior to library screening, sera were absorbed with E. coli DH5α whole cells in order to prevent non-specific selection of clones by E. coli-specific antibodies and thus facilitate the selection of clones by antibodies recognizing K. pneumoniae epitopes. Subsequently, IgG were purified from pooled sera and tested for immune reactivity toward K. pneumoniae MGH 78578 proteins using whole bacterial extracts in western blot (data not shown).
Selection of K. pneumoniae antigens by human sera: the ANTIGENome
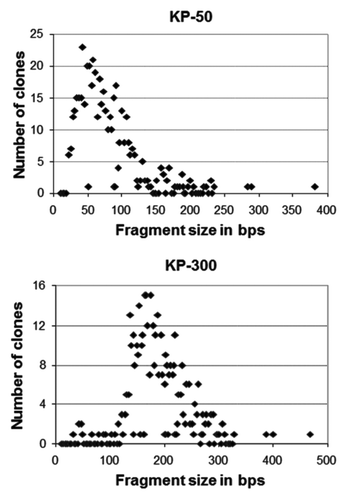
To express Klebsiella-derived epitope-bearing fragments on the surface of E. coli, two different genomic libraries were generated from the K. pneumoniae strain MGH 78578 using two platform proteins for surface expression, LamB and FhuA. In order to generate the LamB library, genomic DNA was digested with DNase I into approximately 50 bp-long fragments. By contrast, the FhuA library was generated by sonication of DNA resulting in app. 150–300 bp-long fragments. The fragments were first cloned into a frame selection vector as described previously.Citation18 To assess the quality of the libraries, approximately 500 randomly chosen clones were sequenced from each of the frame selection libraries. Bioinformatics analysis showed that the actual insert length was close to the intended size (average length, LamB 77 bps and FhuA 187 bps) and that the majority of clones fell within a narrow size range (). After quality control of the frame selection libraries, inserts were transferred into the display vectors, creating bacterial surface display libraries that present epitope-bearing fragments from K. pneumoniae on the surface of E. coli.Citation18
Figure 2. Fragment size distribution of genomic libraries in the LamB (KP-50) and FhuA platform (KP-300). Approximately 500 clones were sequenced from each library and the precise length of the inserts was determined. The numbers of clones with distinct sizes are plotted for each library.
In order to identify immunoreactive Klebsiella proteins, both LamB and FhuA bacterial surface display libraries were incubated with human IgG pools as described previously.Citation18 Using streptavidin-coated microbeads, clones carrying inserts recognized by antibodies were separated by magnetic-activated cell sorting (MACS). Selected E. coli clones were analyzed by western blotting to confirm their antigenicity (data not shown) and subsequently, approximately 8,000 clones were picked from a total of 8 screens and their inserts were sequenced. The sequences were then mapped to the published genome of K. pneumoniae strain MGH 78578 (NCBI accession number CP000647). This resulted in the identification of 169 sequences which were identified at least twice. This comprehensive antigen identification approach has led to the discovery of 74 annotated and 95 non-annotated genes encoding potential antigens of K. pneumoniae. A large number of the identified antigens belong to the category of surface exposed or secreted proteins, as would be expected from an extracellular pathogen. Besides the antigens derived from annotated ORFs, a considerable number of potential antigenic peptides could be traced back to non-annotated regions of the genome. These alternative reading frames (ARFs) or complementary reading frames (CRFs) have a variable and mostly short length ranging from 25 to 823 amino acids and did not show any significant homology to known proteins. At this point it can only be speculated that these antigenic peptides delineate either novel ORFs which have not been assigned a function by the genomic annotation process or were identified due to a cross reaction of serum antibodies to other epitopes (Table S1). The most antigenic among all 169 selected candidates, identified 83 times, was the non-annotated antigen KPA-03. The second most frequently recognized candidate, with 79 hits, was the non-annotated antigen KPC-03. The most frequently selected annotated ORF was the glutamine ABC transporter substrate-binding protein (KP-64). In total, annotated (74 candidates) and non-annotated proteins (95) were selected at similar frequency, indicating the potential of novel ORFs to be identified in the Klebsiella ANTIGENome. When analyzing the selected antigens with known function, membrane proteins and secreted proteins formed a considerable group of 23 and 13 antigens, respectively. Among them the protein FepA (KP-66),Citation16 already known to be potential vaccine candidate. It is also worth mentioning that we did not identify any outer membrane protein (OMP),Citation16 or fimbriae protein, like type 3 fimbriae,Citation23 in our screens.
Selection of 26 vaccine candidates based on in vitro validation assays
In order to select the most promising candidate antigens for further evaluation in animal models, we performed peptide ELISA with human sera and a gene conservation analysis. In addition, we analyzed all antigens by bioinformatics for homology with human proteins and for novelty in regard to published homologs proteins. As a first step we investigated the presence of the antigen encoding genes in a collection of human pathogenic Klebsiella strains. For the ARFs and CRFs, candidates were only included in the evaluation process if they were identified more than 30 times or the length of the encoded peptide was predicted to be longer than 100 amino acids. A panel of 46 different Klebsiella isolates, comprising the most relevant major O- and K-types, was tested by PCR for the presence of the respective genes. Of the 103 selected antigens analyzed, 60 were detected in more than 85% of the strains tested, of which 21 genes were identified in all strains (Tables S1 and S3).
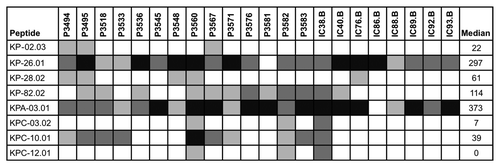
The identification of epitope-bearing fragments in the bacterial surface display libraries is dependent on the amount and affinity of the specific antibodies in the applied serum pools. In order to assess the extent of antigenicity, the presence of antibodies in individual human sera specific for the identified epitope-bearing fragments was determined by peptide ELISA. The human sera used for this analysis were mainly those included in the six serum pools applied for the identification of antigens by bacterial surface display. In total, 214 overlapping synthetic biotin-labeled peptides of approximately 25 to 30 amino acids in length covering the epitope-bearing fragments (clone regions) representing the selected immunogenic epitopes in 74 ORFs and 29 non-annotated ORFs, were used to coat streptavidin ELISA plates. Many of the peptides were confirmed to be antigenic (Table S2). For some of the antigens, it was observed that peptides representing different regions showed various degree of reactivity. As an example, the results for four ORFs and four non-annotated antigens which were identified representing antigens of low, intermediate and high frequency (hits from 2 to 79) are shown in . Peptides from some antigens (KP-26, KP-82 and KPA-03) were highly reactive with the human sera, on the other hand some peptides showed only low or no reactivity with the human sera, such as KP-02 and KPC-12. Overall, the frequency of identification did not correlate with the median ELISA titer. One explanation for the selection of such antigens by our screens despite the lack of ELISA reactivity might be the possibility that antibodies against these antigens recognized conformational rather than linear epitopes.
Figure 3. Peptide ELISA with human sera. Reactivity of human sera against KP-02, KP-26, KP-28, KP-82, KPA-03, KPC-03, KPC-10 and KPC-12 are shown. Reactivity of the most reactive peptides per antigen are shown. Coding of reactivity: < 100 ELISA units (white), 100 - 199 ELISA units (light gray), 200 - 399 ELISA units (dark gray), > 400 ELISA units (black).
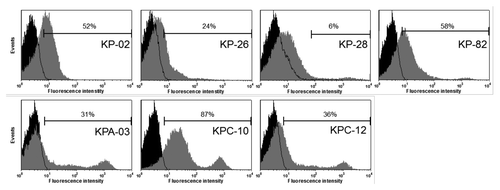
One additional important criterion for the selection of a vaccine candidate is its surface exposure and thus accessibility for binding by human antibodies. In order to address this issue, immune sera against epitope-bearing fragments were generated in mice by immunization with total bacterial lysates prepared from MACS screen-selected clones. Sera against 103 antigens were successfully generated and tested by flow cytometry with live in vitro grown K. pneumoniae strain A5054 and Friedländer 204 (K- mutant) cells. These analyses indicated surface exposure of 77 antigens (Table S1), from which 16 showed prominent shifts for over 35% of the cells. Among these surface exposed antigens were some non-annotated antigens (e.g., KPA-03 and KPC-10) as shown in , the known immunoreactive protein KP-66 (FepA), membrane protein KP-78 (LamB) and membrane-associated protein KP-26 (TonB). However, it should be kept in mind that some surface exposed antigens present in the Klebsiella ANTIGENome were potentially missed by our surface exposure analysis, because they were not expressed in in vitro grown cells, or because the selected peptides may not be readily accessible on the surface.
Figure 4. Surface staining of K. pneumoniae A5054. Cells were stained with immune sera from mice immunized with bacterial lysates derived from clones expressing epitope-bearing fragments fused to the respective surface-located platform protein. Data for four annotated ORFs and three non-annotated ORFs are shown as examples.
Based on the results of the different in vitro validation steps (prevalence of the antigen in the tested Klebsiella strains, their high reactivity with human sera and/or confirmation to be surface located), 22 of the 74 annotated antigens were selected for further studies. In addition, we also included four of the non-annotated antigens (KPA-03, KPC-10, KPC-12 and KPC-14, ). These vaccine candidates were cloned into the pET28b vector for expression as recombinant proteins. Whenever it was possible, full-length proteins were expressed; however, ancillary sequences encoding, e.g., signal peptides or lipoprotein signal peptides were not included. Some of the genes were cloned in several smaller fragments due to their large size, which was predicted or shown to hinder efficient expression of the encoded full-length protein. For 24 antigens, with the exception of antigens KP-56 and KP-58, expression constructs could be generated and recombinant proteins were expressed and purified successfully.
Table 1. List of proteins selected by validation analysis
Eight ANTIGENome-derived Klebsiella antigens provide protection against Klebsiella infection in a mouse sepsis model
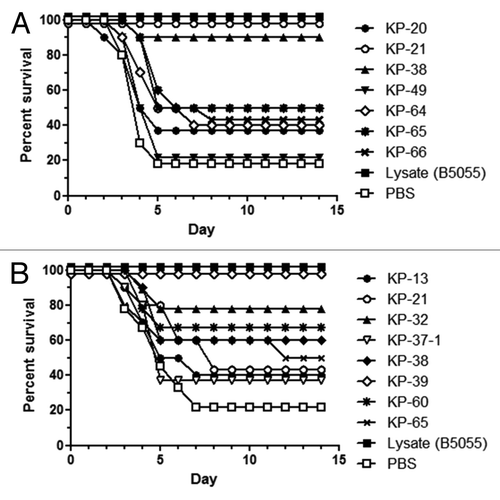
To determine whether the 24 selected Klebsiella antigens elicit protective immune responses, and thus qualify as candidates with vaccine potential, immunization-challenge studies needed to be conducted in mice. K. pneumoniae strain B5055 was selected as the challenge strain for active and passive immunization experiments as this strain has been used successfully to establish sepsis and pneumonia in mice.Citation24-Citation26 The LD90 dose for an intraperitoneal challenge model of K. pneumoniae infection was established in CD-1 mice. Animals were immunized subcutaneously with the recombinant antigens formulated with CFA-IFA as adjuvant (up to six individual experiments; 10 mice/group). These studies showed that eight Klebsiella proteins, namely, KP-13, KP-21, KP-32, KP-37, KP-38, KP-39, KP-60, and KP-65, were capable of inducing significant protection against Klebsiella infection in a CD-1 mouse sepsis model. The protection levels ranged from 20% above the negative control to a maximum of 100% survival in individual experiments. Protection was evaluated based on survival rates up to day 14 post-challenge and statistically significant differences are based on Log-Rank (Mantel-Cox) analyses of Kaplan-Meier plots. One representative experiment is shown in with three antigens, KP65 (p-value = 0.0399), KP38 (p-value = 0.0018) and KP-21 (p-value = 0.0003), which provided statistically significant protection. In addition, we also evaluated these antigens using aluminum hydroxide [Al(OH)3] as adjuvant (two independent experiments). In these experiments, the protection levels ranged from 20% above the negative control to a maximum of 100% survival, with two antigens, KP32 (p-value = 0.0245) and KP39 (p-value = 0.0004) showing a statistical significant level of protection (). Furthermore, we selected five out of the eight proteins (KP-13, KP-21, KP-38, KP-39 and KP-60) for generation of rabbit hyper-immune serum to perform passive protection experiments. CD-1 mice were immunized with 150 µL of individual hyper-immune sera and challenged intraperitoneally 3 h later. Intermediate but no significant protection was observed only for KP-60 (60% above the control level) in one experiment (data not shown).
Figure 5. Protection conferred by active immunization with selected K. pneumoniae antigens in a mouse lethality model. CD-1 mice were immunized subcutaneously with recombinant antigens and challenged with K. pneumoniae strain B5055. Survival was monitored for 14 d post-challenge and presented as Kaplan–Meier survival curves. Mice were immunized subcutaneously with 50 µg recombinant protein with (A) CFA/IFA as adjuvant or (B) with aluminum hydroxide. Mice were challenged intraperitoneally with 103 CFU K. pneumoniae B5055 and the numbers of surviving mice were plotted as a percentage of total mice.
The eight selected vaccine candidates have not been characterized previously in more detail and only putative functions were assigned by genome annotation. KP13 (KPN_02645) was annotated as glycerophosphodiester phosphodiesterase. KP21 (KPN_03030) has a putative function as recombination regulator RecX and the protein might be a regulator of RecA activity by interaction with the RecA protein or filament. KP32 (KPN_03857) shows similarities to oxidoreductase with a FAD/NAD(P)-binding domain. KP37 (KPN_03752) is assigned as nitrite reductase [NAD(P)H] subunit. KP38 (KPN_02850) was described as a α2-macroglobulin (yfhM) involved in colonization and YfhM homologs in bacteria are always paired with a pbpC gene encoding a very efficient peptidoglycan glycosyltransferase. YfhM and PBP1C may act as a defense and repair mechanism to protect the cell from host defenses during colonization. KP39 (KPN_02890) is predicted to be located in the cytoplasm and involved in DNA repair. KP60 (KPN_00840) display similarities to glutamine ABC transporter periplasmic protein. Bacterial high affinity transport systems are involved in active transport of solutes across the cytoplasmic membrane. KP65 (KPN_00667) is annotated as penicillin-binding protein 2, which is a protein located in either the cytoplasm or associated with the membrane.
Genomic conservation of Klebsiella vaccine candidates
The goal of the present study was the selection of conserved protective vaccine candidates. We thus extended the gene distribution study beyond the detection of the genes via PCR (Table S4) and evaluated the sequence conservation of the eight protective proteins in a collection of representative isolates of Klebsiella (Table S3). The 8 genes displayed a level of protein sequence identity larger than 92% in all strains analyzed, with the exception of KP-21 where the protein identity was in some strains as low as 83%. Partial sequences were obtained for KP-38, as the nucleotide sequence encoding the very C terminus was not successfully amplified, but all analyzed sequences comprised the corresponding amino acid sequence of the recombinant KP-38 fragment used for immunization-challenge studies.
The obtained data confirmed the high level of sequence conservation of all 8 proteins by pairwise alignments (KP-13, > 95.8%; KP-21, > 83.3%; KP-32, 92.7%; KP-37, > 99.4%; KP-38, > 98.2%; KP-39, > 93.1%; KP-60, > 94.4%; KP-65, > 98.3%).
Discussion
Development of vaccines to prevent K. pneumoniae infection is driven by the fact that nosocomial Klebsiella infections continue to be a heavy burden on the economy and on the life expectancy of patients in developed and developing countries. K. pneumoniae is among the 10 most common pathogens which account for 84% of any healthcare-associated infections.Citation27 While K. pneumoniae can cause bacterial pneumonia leading to extensive lung damage, the most common site of infection is the urinary tract, with Klebsiella spp accounting for approximately 6–17% of all nosocomial urinary tract infections (UTI).Citation27,Citation28 Klebsiella ranks second only to E. coli as the cause of bacteremia due to biliary tract infection (BTI).Citation29 It is also frequently implicated as a cause of wound infections, particularly in immunocompromised individuals and is in intensive care units-acquired pneumonia among P. aeruginosa, S. aureus, E. coli and Enterobacter spp the most frequently isolated microorganism.Citation22
Vaccine development for Klebsiella has so far focused on the five main classes of epitopes identified for Klebsiella pathogenic mechanisms and which are recognized by the immune system: capsule, LPS, siderophores, adhesins and exotoxins.Citation4 The most commonly recognized antigenic structures to induce both humoral and cellular immune response are polysaccharides (LPS and CPS). The production of a 24-valent CPS vaccine showed excellent antibody response in traumatic patients,Citation8 but the multivalent vaccine only covered 70% of all Klebsiella bacteremia isolates, whereas the small number of different O-types,Citation30 is an advantage for LPS as a vaccine candidate and it has been shown recently that O-antigens are surface exposed in strains expressing certain capsular polysaccharides.Citation31 An O1-antigen specific antibody was able to protect against capsular and non-capsular Klebsiella causing sepsis,Citation32 and a monoclonal antibody specific for the O1-antigen provided protection in a mouse model of lethal endotoxemia.Citation33
Protein candidates on the other hand, if conserved, accessible and expressed by most strains of K. pneumoniae would be an appropriate alternative to polysaccharides. Protein based vaccines were focusing on immunogenic proteins like extracellular toxins or cell surface proteins, such as outer membrane proteins (Omp) and fimbriae proteins.Citation23,Citation34 Type 1 and 3 fimbriae are an essential virulence factor in K. pneumoniae UTI, although expression of type 3 fimbriae had no influence on pathogenicity in an UTI animal model.Citation35,Citation36 Type 3 fimbriae-mediated immunity could be demonstrated in a murine model of respiratory disease.Citation23 Rats immunized with a heat stable enterotoxin from K. pneumoniae were protected against active challenge and showed immunological cross-reactivity with E. coli enterotoxin.Citation37 The individual antigens have been selected by various technologies. OmpA has been identified by a novel multi-screening signature-tagged mutagenesis assay,Citation38 the antigen FepA was selected by a novel in vivo expression technology,Citation39 whereas the antigens OmpK36, OmpK17, OmpW, FepA and Colicin I receptor were identified by a immunoproteome based approach.Citation16 We embarked on a genome-wide selection of antigens, employing sera from patients with skin and soft tissue infection, pneumonia, septicemia, intra-abdominal infection and urinary tract infection, as well as from healthy individuals exposed to, but not colonized by K. pneumoniae. In order to avoid any potential shortcomings by the lack of in vitro expression or proper bioinformatics annotation of protein function, we applied the ANTIGENome technology,Citation18,Citation19,Citation40 to select novel protective Klebsiella vaccine candidates on a genomic scale. As a result, we identified 169 antigenic proteins from K. pneumoniae, including the already known protective antigen FepA. By applying several in vitro assays and a murine infection model, we identified eight novel proteins capable of providing protection against K. pneumoniae infection by active immunization (), and confirming the validity of the technology to identify relevant candidates.
Our preliminary data on passively administrated rabbit sera showed no protection in a murine infection model, however passive transfer of polyclonal rabbit sera raised against CPS-antigens protected mice against lethal infections,Citation41 and the protection against Klebsiella infection mediated by anti-lipopolysaccharide antibodies is dependent on the production of the anti-inflammatory cytokine IL-10.Citation42 The host defense against Klebsiella by the adaptive immune system is not only dependent on the humoral response but also on the cellular immunity.Citation43 The recent discovery of the T-cell cytokine IL-17 has added to the understanding of host defense mechanisms that IL-17 servers as a link between adaptive and innate immunity that extends beyond protection against extracellular bacterial and fungal pathogens.Citation44,Citation45
All eight vaccine candidates described in this study are highly conserved not only in the 7 published Klebsiella genomes, but also in 46 clinical isolates that we analyzed. Thus, it can be assumed that an immune response against any of these proteins will be able to recognize pathogenic strains if the respective protein is surface accessible during the infection. The observed high level of sequence conservation further argues that these proteins provide an important function to the pathogen and development of escape mutants are not very likely. Despite the present lack of understanding of their precise role in Klebsiella virulence, our studies clearly showed that all 8 candidates could individually induce protection in mice upon active immunization. As K. pneumoniae has evolved different mechanisms for evasion of the immune system, thus, inclusion of more than one protein in a vaccine will increase the likelihood of inducing a broad protective immune responses and prevention of disease.
The eight identified protective antigens from K. pneumoniae are highly conserved in a representative selection of Klebsiella serotypes, and several of the identified candidates were shown or predicted to be involved in the pathogenesis of Klebsiella. This makes them suitable candidates for a K. pneumoniae vaccine, and we therefore currently evaluate and select combinations of proteins to achieve a high level of protection.
Material and Methods
Klebsiella strains and culture conditions
K. pneumoniae strain MGH 78578, a clinical isolate from sputum, used for generation of bacterial surface display libraries and as a source for cloning of antigens was obtained from the American Type Culture Collection (ATCC number 700721). K. pneumoniae strains A5054, B5055, Friedländer 204, Mich 61, 708, and the capsule negative mutant Mich 61 and 708 were obtained from Statens Serum Institut (SSI), Denmark. The Klebsiella strains used for the gene distribution studies were mainly clinical isolates of K. pneumoniae, K. ozeanae, K. planticola and K. oxytoca and obtained from SSI. Bacteria were grown at 37°C in Nutrient Broth (Difco 234000). For preparation of total bacterial lysate, cells were lysed by repeated freeze-thaw cycles, followed by incubation on dry ice/ethanol-mixture until frozen for 1 min and thawing at 37°C for 5 min. The freeze-thaw cycle was repeated 3 times, followed by sonication. After centrifugation at 4,000 rpm for 15 min at 4°C, the supernatants containing soluble material were collected, whereas the pellets containing cell debris and insoluble cell material were discarded.
Generation of bacterial surface display libraries in E. coli and magnetic cell sorting
Generation of bacterial surface display libraries in E. coli and subsequent identification of the Klebsiella ANTIGENome was performed as described previously.Citation18,Citation20
Human serum samples
A collection of 100 human sera was obtained from patients with medical diagnosis of Klebsiella infection in the convalescent phase. The patient serum samples were obtained from Franz-Josef Schmitz (Klinikum Minden, Germany). The disease symptoms recorded and medical diagnosis of these patients included skin and soft tissue infection, pneumonia, septicemia, intra-abdominal infection and urinary tract infection. In addition, a collection of 89 human sera was obtained from healthy individuals. All human patient serum samples, as well as those from healthy individuals, were analyzed by ELISA using whole cell lysates of K. pneumoniae MGH 78578, Mich 61, 708, and the capsule negative mutants of Mich 61 and 708. Disease specific and healthy individuals’ sera with high titer were pooled and IgG isolated as described elsewhere.Citation21
Gene distribution analysis
Oligonucleotide pairs for all identified ORFs and selected ARFs (alternative reading frames) and CRFs (complementary reading frames), were designed using the online tool Primer3 (http://frodo.wi.mit.edu). Genomic DNA from 46 Klebsiella spp that differ in the two major types of antigens on the cell surface (K- and O-type) was used as template for PCR analysis and DNA products were subsequently visualized by electrophoresis.
Gene conversation of Klebsiella antigens
In order to determine the sequence of an antigen from diverse Klebsiella spp, PCR was performed with primers specific for the genes of interest, as described above. Klebsiella spp used for these analyses are shown in Table S2. Oligonucleotide sequences as primers for PCR were designed for the selected antigens in order to be able to amplify the complete gene. The sequences of the oligonucleotides are listed in Table S3. Genomic DNA of all Klebsiella spp was prepared as described above. PCR was performed as described above, unless conditions had to be adapted for individual primer pairs.
Peptide ELISA
Peptide ELISA was performed using a Gemini 160 ELISA robot (TECAN, Austria) as previously described.Citation21 Results are expressed as ELISA units [A405(sample-blank) × 1000].
Surface staining of Klebsiella pneumonia cells
Immune sera were generated in NMRI mice (Harlan, Italy) by immunization with pools of up to five total bacterial lysates (corresponding to 5 × 107 cells) prepared from screen-selected clones expressing epitope-bearing fragments. Approximately 5x105 bacteria in 100 µL HBSS medium were incubated with 5 µL of heat inactivated mouse sera for 1 h on ice before detection with PE-conjugated goat anti-mouse IgG (H+L; Beckman Coulter, Austria) antibodies. After fixation with 2% paraformaldehyde, surface staining was detected by flow cytometry (Cytomics FC500; Beckman Coulter), and data were analyzed using the analysis software FCS Express (De Novo Software, USA).
Cloning and expression of recombinant proteins
The gene fragments of interest were amplified from K. pneumoniae strain MGH 78578 genomic DNA by PCR and cloned into the pET28b(+) vector (Novagen, USA). Proteins were expressed in BL21-CodonPlus (DE3)-RIPL cells (Life Technologies, USA) with a His-tag at the C-terminus. A single colony was inoculated in a 5 mL LB supplemented with 50 µg/mL kanamycin, incubated overnight, and subsequently expanded to a volume of 500 mL or up to 5 L. Cells were induced with 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside) at an OD600 of 0.8, and growth continued for 3–4 h, thereafter cells were harvested by centrifugation. Lysis was performed by a combination of the freeze-thaw method and treatment with BugBuster (Novagen). The lysate was separated by centrifugation at 12,000 g for 30 min into soluble (supernatant) and insoluble (pellet) fractions. For the proteins that were expressed in the soluble fraction, purification of the protein was performed by binding the supernatant to Ni-Sepharose column (Ni Sepharose 6 Fast Flow; GE Healthcare, United Kingdom) as recommended by the manufacturer. For proteins in the insoluble fraction, the pellet was washed 3 times with 50 mM TRIS-HCl, pH 8.0, 100 mM NaCl, 0.5% Triton X-100 and then solubilized in a suitable buffer containing 8 M urea. The purification was performed under denaturing conditions in buffer containing 8 M urea. The eluate was concentrated and dialyzed to remove all urea in a gradual and stepwise manner.
Animal challenge studies
All animal experiments were conducted in accordance with Austrian law (BGB1 Nr. 501/1989) and approved by “Magistratsabteilung 58.” Outbred CD-1 mice (Harlan, Italy) were used for the immunological studies. Prior to each challenge, groups of five 8 week-old female mice were bled via the tail vein and pre-immune sera were prepared and pooled. For active immunization (subcutaneous route), 50 µg of recombinant proteins formulated with either Complete Freund’s adjuvant (CFA) or 2% aluminum hydroxide [Al(OH)3] as adjuvant were injected into CD-1 mice. On days 14 and 28, mice were boosted with the same amount of protein and adjuvant, [except that Incomplete Freund’s adjuvant (IFA) was used rather than CFA]. Mice immunized with K. pneumoniae B5055 lysate served as a positive control, while mice immunized with adjuvant only, served as a negative control. Antibody titers were measured at day 35 by ELISA using the respective recombinant proteins. For passive immunization [intraperitoneal route (IP)], 150 µL of hyper-immune rabbit sera (Charles River, Germany) raised against individual, K. pneumoniae recombinant protein antigens was injected into CD-1 mice, three hours prior to IP bacterial challenge. Antibody titers of the sera used for immunization were measured using the respective recombinant proteins.
Bacterial challenge: Freshly grown K. pneumoniae strain B5055 was used. In order to determine the viable cell numbers present in the bacterial inoculum, CFU were determined by plating dilutions of the inoculum onto blood agar plates. 103 CFU were applied IP. Protection conferred by immunization was measured using a bacteremia/sepsis model in which survival rates were followed for 2 weeks post-challenge, and survival was expressed as a percentage of the total number of animals (10 mice/group).
ELISA
ELISA plates (Maxisorb, Nunc, Denmark) were coated with 5–10 µg/mL total protein diluted in coating buffer (0.1 M sodium carbonate pH 9.2). Two dilutions of sera (1:1,000 and 1:5,000) were made in PBS-BSA. Highly specific horseradish peroxidase (HRP)-conjugated anti-human IgG secondary antibodies (Southern Biotech, USA) were used according to the manufacturers' recommendations (dilution: 1:1,000). Alternatively, horseradish peroxidase (HRP)-conjugated anti-mouse IgG was used for mouse sera. Antigen-antibody complexes were quantified by measuring the conversion of the substrate (ABTS) to colored product based on A405nm readings by automatic ELISA reader (TECAN SUNRISE).
Immunoblotting
Binding of post challenge sera to recombinant proteins was analyzed by immunoblotting. Briefly, 1 µg of each recombinant protein was separated by SDS-PAGE under reducing conditions on 4–12% Tris-Glycine ZOOM gels (Life Technologies). Separated proteins were transferred onto nitrocellulose membrane using the iBlot® Dry blotting system (Life Technologies). After overnight blocking in 5% milk, post-challenge sera (pooled from five mice) were added at 1:1000 dilution and polyclonal rabbit anti-mouse IgG conjugated to HRP (Dako, Denmark) was used for detection. The immunoblots were visualized with Amersham ECL PlusTM western blotting detection reagents (GE Healthcare) and Kodak BioMax films (Kodak, USA).
Additional material
Download Zip (530.1 KB)Acknowledgments
The authors wish to express their gratefulness to Birgit Noiges, Christine Triska, Ulrike Stierschneider, Christina Satke, Thomas Cipps and Dieter Gelbmann for their technical support. Eszter Nagy for her dedicated support throughout this project.
Submitted
10/18/12
Accepted
10/27/12
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- Stamm WE, Weinstein RA, Mannheim W. Comparison of endemic and epidemic nosocomial infection. Nosocomial infection. In: Dixon R E, ed. Nosocomial infections. Atlanta, Ga: Yorke Medical Books; 1981:9-13.
- Zaidi AK, Huskins WC, Thaver D, Bhutta ZA, Abbas Z, Goldmann DA. Hospital-acquired neonatal infections in developing countries. Lancet 2005; 365:1175 - 88; http://dx.doi.org/10.1016/S0140-6736(05)71881-X; PMID: 15794973
- Wisplinghoff H, Bischoff T, Tallent SM, Seifert H, Wenzel RP, Edmond MB. Nosocomial bloodstream infections in US hospitals: analysis of 24,179 cases from a prospective nationwide surveillance study. Clin Infect Dis 2004; 39:309 - 17; http://dx.doi.org/10.1086/421946; PMID: 15306996
- Podschun R, Ullmann U. Klebsiella spp. as nosocomial pathogens: epidemiology, taxonomy, typing methods, and pathogenicity factors. Clin Microbiol Rev 1998; 11:589 - 603; PMID: 9767057
- Wu LT, Hung SW, Chuang YC, Chen HE, Jones RN, Yu WL. Identification of a novel cephalosporinase (DHA-3) in Klebsiella pneumoniae isolated in Taiwan. Clin Microbiol Infect 2005; 11:893 - 7; http://dx.doi.org/10.1111/j.1469-0691.2005.01252.x; PMID: 16216104
- Hawkey P, Finch R. Tigecycline: in-vitro performance as a predictor of clinical efficacy. Clin Microbiol Infect 2007; 13:354 - 62; http://dx.doi.org/10.1111/j.1469-0691.2006.01621.x; PMID: 17359318
- Ahmad TA, El-Sayed LH, Haroun M, Hussein AA, El Ashry SH. Development of immunization trials against Klebsiella pneumoniae. Vaccine 2012; 30:2411 - 20; http://dx.doi.org/10.1016/j.vaccine.2011.11.027; PMID: 22100884
- Campbell WN, Hendrix E, Cryz S Jr., Cross AS. Immunogenicity of a 24-valent Klebsiella capsular polysaccharide vaccine and an eight-valent Pseudomonas O-polysaccharide conjugate vaccine administered to victims of acute trauma. Clin Infect Dis 1996; 23:179 - 81; http://dx.doi.org/10.1093/clinids/23.1.179; PMID: 8816151
- Chhibber S, Bajaj J. Polysaccharide-iron-regulated cell surface protein conjugate vaccine: its role in protection against Klebsiella pneumoniae-induced lobar pneumonia. Vaccine 1995; 13:179 - 84; http://dx.doi.org/10.1016/0264-410X(95)93133-T; PMID: 7625113
- Edelman R, Taylor DN, Wasserman SS, McClain JB, Cross AS, Sadoff JC, et al. Phase 1 trial of a 24-valent Klebsiella capsular polysaccharide vaccine and an eight-valent Pseudomonas O-polysaccharide conjugate vaccine administered simultaneously. Vaccine 1994; 12:1288 - 94; http://dx.doi.org/10.1016/S0264-410X(94)80054-4; PMID: 7856293
- Cryz SJ Jr., Mortimer PM, Mansfield V, Germanier R. Seroepidemiology of Klebsiella bacteremic isolates and implications for vaccine development. J Clin Microbiol 1986; 23:687 - 90; PMID: 3517058
- Yadav V, Sharma S, Harjai K, Mohan H, Chhibber S. Lipopolysaccharide-mediated protection against Klebsiella pneumoniae-induced lobar pneumonia: intranasal vs. intramuscular route of immunization. Folia Microbiol (Praha) 2005; 50:83 - 6; http://dx.doi.org/10.1007/BF02931298; PMID: 15954538
- Cryz SJ Jr., Fürer E, Germanier R. Safety and immunogenicity of Klebsiella pneumoniae K1 capsular polysaccharide vaccine in humans. J Infect Dis 1985; 151:665 - 71; http://dx.doi.org/10.1093/infdis/151.4.665; PMID: 3882856
- Cryz SJ Jr., Mortimer P, Cross AS, Fürer E, Germanier R. Safety and immunogenicity of a polyvalent Klebsiella capsular polysaccharide vaccine in humans. Vaccine 1986; 4:15 - 20; http://dx.doi.org/10.1016/0264-410X(86)90092-7; PMID: 3962447
- Cryz SJ Jr., Mortimer P, Cross AS, Fürer E, Germanier R. Safety and immunogenicity of a polyvalent Klebsiella capsular polysaccharide vaccine in humans. Vaccine 1986; 4:15 - 20; http://dx.doi.org/10.1016/0264-410X(86)90092-7; PMID: 3962447
- Kurupati P, Teh BK, Kumarasinghe G, Poh CL. Identification of vaccine candidate antigens of an ESBL producing Klebsiella pneumoniae clinical strain by immunoproteome analysis. Proteomics 2006; 6:836 - 44; http://dx.doi.org/10.1002/pmic.200500214; PMID: 16372264
- Kurupati P, Ramachandran NP, Poh CL. Protective efficacy of DNA vaccines encoding outer membrane protein A and OmpK36 of Klebsiella pneumoniae in mice. Clin Vaccine Immunol 2011; 18:82 - 8; http://dx.doi.org/10.1128/CVI.00275-10; PMID: 21048001
- Etz H, Minh DB, Henics T, Dryla A, Winkler B, Triska C, et al. Identification of in vivo expressed vaccine candidate antigens from Staphylococcus aureus. Proc Natl Acad Sci U S A 2002; 99:6573 - 8; http://dx.doi.org/10.1073/pnas.092569199; PMID: 11997460
- Giefing C, Meinke AL, Hanner M, Henics T, Bui MD, Gelbmann D, et al. Discovery of a novel class of highly conserved vaccine antigens using genomic scale antigenic fingerprinting of pneumococcus with human antibodies. J Exp Med 2008; 205:117 - 31; http://dx.doi.org/10.1084/jem.20071168; PMID: 18166586
- Henics T, Winkler B, Pfeifer U, Gill SR, Buschle M, von Gabain A, et al. Small-fragment genomic libraries for the display of putative epitopes from clinically significant pathogens. Biotechniques 2003; 35:196 - 202, 204, 206 passim; PMID: 12866421
- Meinke A, Henics T, Hanner M, Minh DB, Nagy E. Antigenome technology: a novel approach for the selection of bacterial vaccine candidate antigens. Vaccine 2005; 23:2035 - 41; http://dx.doi.org/10.1016/j.vaccine.2005.01.005; PMID: 15755567
- www.ecdc.europa.eu/en/publications/Publications/120215_SUR_HAI_2007.pdf
- Lavender H, Jagnow JJ, Clegg S. Klebsiella pneumoniae type 3 fimbria-mediated immunity to infection in the murine model of respiratory disease. Int J Med Microbiol 2005; 295:153 - 9; http://dx.doi.org/10.1016/j.ijmm.2005.04.001; PMID: 16047414
- Regué M, Hita B, Piqué N, Izquierdo L, Merino S, Fresno S, et al. A gene, uge, is essential for Klebsiella pneumoniae virulence. Infect Immun 2004; 72:54 - 61; http://dx.doi.org/10.1128/IAI.72.1.54-61.2004; PMID: 14688080
- Kuenen JD, van Dijke EE, Hol C, Bootsma HJ, Verhoef J, van Dijk H. Protective effects of orally administered, Klebsiella-containing bacterial lysates in mice. FEMS Immunol Med Microbiol 1994; 8:69 - 75; http://dx.doi.org/10.1111/j.1574-695X.1994.tb00427.x; PMID: 8156053
- Tomás JM, Benedí VJ, Ciurana B, Jofre J. Role of capsule and O antigen in resistance of Klebsiella pneumoniae to serum bactericidal activity. Infect Immun 1986; 54:85 - 9; PMID: 3531020
- Hidron AI, Edwards JR, Patel J, Horan TC, Sievert DM, Pollock DA, et al, National Healthcare Safety Network Team, Participating National Healthcare Safety Network Facilities. NHSN annual update: antimicrobial-resistant pathogens associated with healthcare-associated infections: annual summary of data reported to the National Healthcare Safety Network at the Centers for Disease Control and Prevention, 2006-2007. Infect Control Hosp Epidemiol 2008; 29:996 - 1011; http://dx.doi.org/10.1086/591861; PMID: 18947320
- Ullmann U. [Bacterial infection agents in hospitalized patients]. Zentralbl Bakteriol Mikrobiol Hyg B 1986; 183:103 - 13; PMID: 3107262
- Ortega M, Marco F, Soriano A, Almela M, Martínez JA, López J, et al. Epidemiology and prognostic determinants of bacteraemic biliary tract infection. J Antimicrob Chemother 2012; 67:1508 - 13; http://dx.doi.org/10.1093/jac/dks062; PMID: 22408140
- Hansen DS, Mestre F, Alberti S, Hernández-Allés S, Alvarez D, Doménech-Sánchez A, et al. Klebsiella pneumoniae lipopolysaccharide O typing: revision of prototype strains and O-group distribution among clinical isolates from different sources and countries. J Clin Microbiol 1999; 37:56 - 62; PMID: 9854064
- Tomas JM, Camprubi S, Williams P. Surface exposure of the O-antigen in Klebsiella pneumoniae O1:K1 serotype strains. Microb Pathog 1988; 5:141 - 7; http://dx.doi.org/10.1016/0882-4010(88)90016-2; PMID: 3070259
- Rukavina T, Tícac B, Susa M, Jendrike N, Jonjíc S, Lucin P, et al. Protective effect of antilipopolysaccharide monoclonal antibody in experimental Klebsiella infection. Infect Immun 1997; 65:1754 - 60; PMID: 9125558
- Mandine E, Salles MF, Zalisz R, Guenounou M, Smets P. Murine monoclonal antibodies to Klebsiella pneumoniae protect against lethal endotoxemia and experimental infection with capsulated K. pneumoniae. Infect Immun 1990; 58:2828 - 33; PMID: 1696932
- Goetsch L, Gonzalez A, Plotnicky-Gilquin H, Haeuw JF, Aubry JP, Beck A, et al. Targeting of nasal mucosa-associated antigen-presenting cells in vivo with an outer membrane protein A derived from Klebsiella pneumoniae. Infect Immun 2001; 69:6434 - 44; http://dx.doi.org/10.1128/IAI.69.10.6434-6444.2001; PMID: 11553588
- Struve C, Bojer M, Krogfelt KA. Identification of a conserved chromosomal region encoding Klebsiella pneumoniae type 1 and type 3 fimbriae and assessment of the role of fimbriae in pathogenicity. Infect Immun 2009; 77:5016 - 24; http://dx.doi.org/10.1128/IAI.00585-09; PMID: 19703972
- Struve C, Bojer M, Krogfelt KA. Characterization of Klebsiella pneumoniae type 1 fimbriae by detection of phase variation during colonization and infection and impact on virulence. Infect Immun 2008; 76:4055 - 65; http://dx.doi.org/10.1128/IAI.00494-08; PMID: 18559432
- Klipstein FA, Engert RF, Houghten RA. Immunological properties of purified Klebsiella pneumoniae heat-stable enterotoxin. Infect Immun 1983; 42:838 - 41; PMID: 6358035
- Struve C, Forestier C, Krogfelt KA. Application of a novel multi-screening signature-tagged mutagenesis assay for identification of Klebsiella pneumoniae genes essential in colonization and infection. Microbiology 2003; 149:167 - 76; http://dx.doi.org/10.1099/mic.0.25833-0; PMID: 12576590
- Lai YC, Peng HL, Chang HY. Identification of genes induced in vivo during Klebsiella pneumoniae CG43 infection. Infect Immun 2001; 69:7140 - 5; http://dx.doi.org/10.1128/IAI.69.11.7140-7145.2001; PMID: 11598090
- Meinke A, Storm M, Henics T, Gelbmann D, Prustomersky S, Kovács Z, et al. Composition of the ANTIGENome of Helicobacter pylori defined by human serum antibodies. Vaccine 2009; 27:3251 - 9; http://dx.doi.org/10.1016/j.vaccine.2009.01.066; PMID: 19200834
- Roe EA, Jones RJ, Dyster RE. Passive immunization of mice against Klebsiella aerogenes. Br J Exp Pathol 1986; 67:25 - 32; PMID: 3511939
- Rukavina T, Ticac B, Vasiljev V. IL-10 in antilipopolysaccharide immunity against systemic Klebsiella infections. Mediators Inflamm 2006; 2006:69431; http://dx.doi.org/10.1155/MI/2006/69431; PMID: 17392590
- Zisman DA, Strieter RM, Kunkel SL, Tsai WC, Wilkowski JM, Bucknell KA, et al. Ethanol feeding impairs innate immunity and alters the expression of Th1- and Th2-phenotype cytokines in murine Klebsiella pneumonia. Alcohol Clin Exp Res 1998; 22:621 - 7; http://dx.doi.org/10.1111/j.1530-0277.1998.tb04303.x; PMID: 9622442
- Curtis MM, Way SS. Interleukin-17 in host defence against bacterial, mycobacterial and fungal pathogens. Immunology 2009; 126:177 - 85; http://dx.doi.org/10.1111/j.1365-2567.2008.03017.x; PMID: 19125888
- Peck A, Mellins ED. Precarious balance: Th17 cells in host defense. Infect Immun 2010; 78:32 - 8; http://dx.doi.org/10.1128/IAI.00929-09; PMID: 19901061