Abstract
Staphylococcus aureus is a well-recognized, clinically important cause of nosocomial infections, and as such, a vaccine to prevent S. aureus infections would be an important achievement. A Phase IIB/III study of V710, a vaccine containing iron-regulated surface determinant B (IsdB), demonstrated significant sero-conversion rates in cardiovascular surgery patients following a single pre-surgery immunization. However, the vaccine was not efficacious in preventing bacteremia or deep sternal wound infection post-surgery, thus raising the possibility that IsdB might not be available for immune recognition during infection. The purpose of the work described herein was to evaluate and quantify the naturally occurring anti-IsdB levels at baseline and over time during infection, to understand whether IsdB is expressed during a S. aureus infection in hospitalized non-vaccinated patients. We evaluated baseline and follow-up titers in 3 populations: (1) healthy subjects, (2) hospitalized patients with non-S. aureus infections, and (3) hospitalized patients with S. aureus infections. Baseline anti-IsdB levels generally overlapped between the 3 groups, but were highly variable within each group. In healthy subjects, baseline and follow-up levels were highly correlated (Spearman's rho = 0.93), and the geometric mean fold-rise (GMFR) in anti-IsdB levels between study entry and last value was 0.9-fold (95% confidence interval (CI): 0.8 to 1.0 ; p = 0.09), showing no trend over time. The convalescent GMFR in anti-IsdB levels from baseline was 1.7-fold (95% CI: 1.3 to 2.2, p = 0.0008) during S. aureus infection, significantly different from the 1.0-fold GMFR (95% CI: 0.9–1.2, p = 0.60) in non-S. aureus infection, p = 0.005. Additionally, S. aureus isolates (51) obtained from the hospitalized patient group expressed the IsdB protein in vitro. Collectively, these data suggest that IsdB expression levels rise substantially following infection with S. aureus, but not with other pathogens, and IsdB is likely well-conserved across S. aureus strains.
Introduction
Staphylococcus aureus are Gram-positive cocci that are part of the normal human flora, and are found on the skin, in the nares, and in the throatCitation1. Approximately half of the population are either permanently or transiently colonized by S. aureus in their anterior naresCitation2, and a recent report indicates that colonization rates in the throat could be twice as high as in the naresCitation3. S. aureus infection is associated with immunocompromised populations, due to either immunodeficiency or to breaches in epidermal barriers, such as surgical incisions or indwelling devicesCitation4. The observed increase in resistance of these bacteria to different antibacterial compound classes is of great concern, and is compounded by the increased infection rates in populations not traditionally associated with being at risk for S. aureus infection, including prison inmates, sports teams, and men who have sex with menCitation5,Citation6. S. aureus infection is often associated with significant morbidity and mortality as well as an increased burden on the health-care systemCitation7,. These factors highlight the need for effective prophylaxis and treatment strategies.
Iron-regulated surface determinant B (IsdB) is a member of the Isd locus which is thought to function collectively in iron uptake through binding of hemoglobin and transport of heme into the cell.Citation8 The IsdB antigen is highly conserved and expressed on all isolates examined to date.Citation9 Due to upregulation in iron-limited environments,Citation9,Citation10 IsdB is theoretically available for immune recognition during infection in the human host, where iron is highly sequestered.Citation8,Citation11,Citation12 Expression of IsdB was highly upregulated by essentially 100% of strain Becker grown under in vivo conditions in rats.Citation10 Multiple reports have demonstrated IsdB to be a potential vaccine candidate for the prevention of S. aureus infectionCitation9,Citation13-Citation16 in rodent challenge models. Importantly, enhanced protection from lethal sepsis in rodent models was mediated by both IsdB-specific CD4+ T cells,Citation16 and IsdB-specific mAb.Citation10,Citation13,Citation17-Citation19 The protection afforded by vaccination with IsdB in rodents was lost if the animals were challenged with an IsdB deleted strain,Citation9,Citation16 confirming the specificity of the vaccine. An observed rise in anti-IsdB titers induced by IsdB formulated on Merck aluminum adjuvant (MAA) correlated with protection against lethal S. aureus challenge,Citation9 indicating that antibody titers may be used as a nominal biomarker for vaccine efficacy.
V710, a vaccine containing unadjuvanted, lyophilized IsdB, was tested in Phase I trials and found to be safe and immunogenic after a single immunization.Citation20 A recent Phase IIB/III study of V710 (Protocol 003), demonstrated a 7.5-fold (CI 7.2,7.8) increase in anti-IsdB levels from baseline to day of hospital admission (14–60 days after vaccination) in cardiovascular surgery patients, after a single immunization (clintrials.gov NCT01324440). However, the vaccine did not induce protection against post-surgery bacteremia or deep sternal wound infection. Antibody titers were no different between V710 patients who contracted S. aureus infections vs. those who did not.Citation21 To determine if the vaccine target antigen was conserved in the S. aureus strains from the trial, the isdB gene was analyzed in > 250 isolates, and all contained the gene encoding IsdB. A representative subset was examined for IsdB expression, and expression was confirmed in vitro under iron-restricted conditions (data not shown).
A potential explanation for lack of V710 efficacy would be the lack of expression of IsdB during infection. Although in vivo expression of IsdB has been demonstrated in rodent models,Citation10,Citation12 expression in human hosts during infection has not been investigated. To address this question, an investigation was conducted to determine if antibody titers to IsdB increase during the course of S. aureus infection in non-vaccinated patients. Hospitalized patients, including surgical patients, were evaluated for increased titers to IsdB after confirmed S. aureus infection, compared with their baseline titers. Control hospitalized patients, including surgical patients, who had infections with pathogens other than S. aureus, were also evaluated. Lastly, as a comparison to the surgical patients, IsdB titers were monitored in a healthy group of individuals not known to have contracted a S. aureus infection, nor to be at risk for infection.
Results
Study group characteristics
Study group characteristics are shown in , and risk factors for S. aureus infection for the hospital cases and hospital controls are shown in . Of note, the majority of the hospitalized subjects had surgery: 85% of the hospital S. aureus cases (34% prior to hospital admission) and 48% of the hospital controls (10% prior to hospital admission).
Table 1. Demographic characteristics of the study populations.
Table 2. Clinical characteristics of hospitalized patients with S. aureus or other infections
Characterization of S. aureus infection in hospital S. aureus cases
Pathogens were identified from blood (40), cerebrospinal fluid (9), surgical tissue (22), synovial fluid (17), sputum (9), or other sites (5) during routine clinical care. Of the 52 S. aureus isolates, 51 were spa typed; a single isolate failed to grow sufficiently in culture for spa determination. Of the 51 S. aureus isolates typed, 16 (31%) were methicillin-resistant S. aureus (MRSA). The MRSA isolates all carried the staphylococcal chromosome cassette (SCC) mec type IV. The collection had strains from widely divergent spa types, including 2 new spa types, with strain clusters (identical spa type) of up to 9 isolates. The cluster identified in 9 cases, spa type 382 (SCC mec IV), was an MRSA strain, and may have been an outbreak strain in the hospital. Detailed spa information is displayed in Table S1. There was a wide diversity of S. aureus strains in the study, based on the widely divergent spa types obtained. All 51 S. aureus isolates in this study expressed IsdB antigen in vitro, as demonstrated via western blot (as described in the Methods section) with an IsdB-specific monoclonal antibody (data not shown).
Comparison of IsdB-specific IgG levels across study populations at baseline
The baseline anti-IsdB levels generally overlapped between the 3 groups, but were highly variable within each group. (). The geometric mean titer concentrations (GMCs) ranged from 19.6 to 37.2 µg/mL, with overlapping 95% confidence intervals between the three groups. Although hospital S. aureus cases had the largest GMC, there did not appear to be any relation between risk groups and baseline anti-IsdB levels (data not shown).
Table 3. IsdB-specific IgG Levels at study entry.
Longitudinal assessment of IsdB-specific IgG levels in study populations
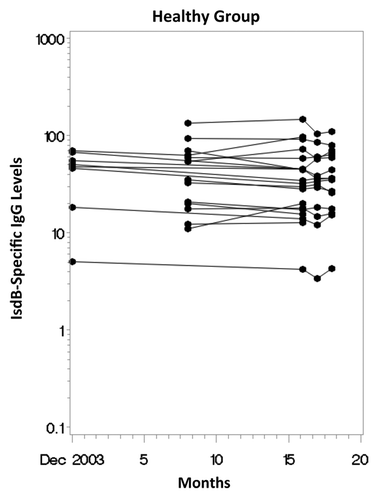
There was a broad range of IsdB-specific IgG levels among adults in the Healthy group at baseline and over study follow-up (). The GMC of the maximum titers was 37.8 μg/mL (range: 5.0 μg/mL to 146.8 μg/mL) and the GMC of the minimum titers was 27.7 μg/mL (range: 3.4 μg/mL to 103.7 μg/mL). There was strong correlation between anti-IsdB levels at study entry and at follow-up (Spearman correlation coefficient: rho = 0.93; p < 0.0001) and between minimum and maximum anti-IsdB levels (rho = 0.95; p < 0.0001). The geometric mean fold-rise(GMFR) in anti-IsdB levels between study entry and last value was 0.9 (95% confidence interval: 0.8 to 1.0 ; p = 0.09), with little change over time in antibody levels for individual subjects.
Figure 1. Longitudinal scatterplot of anti-IsdB antibody levels (ug/mL) over time in the Healthy Group.
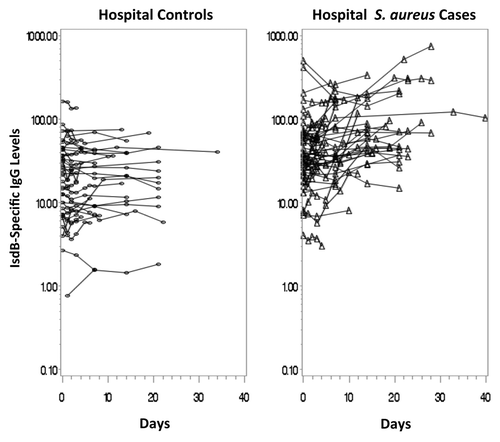
Of the 52 hospital S. aureus cases, 45 had sera available from multiple time-points (median number of samples: 4, range: 2 to 8) with a mean time for final follow-up of 15.3 days post diagnosis (range 2 to 40 days). Of the 50 hospital controls, 39 had sera from multiple time-points (median number of samples: 4, range: 2 to 8), and the mean time for final follow-up was 11.6 days post-diagnosis (range: 2 to 34 days). The mean time of follow-up was slightly longer for the hospital S. aureus cases compared with the hospital controls (p = 0.05). displays a longitudinal scatterplot of anti-IsdB levels per subject over time. The GMC for the hospital S. aureus cases rose from 40.0 µg /mL at the first time-point to 67.4 µg /mL at the end of the study. The GMFR in anti-IsdB titers from study entry to last value was 1.7 (95% CI: 1.3 to 2.2, p = 0.0008). In contrast to the levels in hospital S. aureus cases, there was no rise in levels over time in the hospital controls. The baseline and follow-up GMCs were 17.7 µg /mL and 18.4 µg /mL, respectively, and the GMFR in anti-IsdB titers from study entry to last value was 1.0 (95% CI: 0.9 to 1.2, p = 0.60). There was a significant difference in the rise in levels over time between the hospital S. aureus cases and the hospital controls (p = 0.005). In addition, 16% of the hospital S. aureus cases had final antibody levels greater than 2 times the standard deviation above the geometric mean of the study entry levels for the hospital controls, a proxy for pre-infection levels, compared with 0% of the hospital controls (p = 0.01).
Figure 2. Longitudinal scatterplot of Anti-IsdB antibody levels (ug/mL) over time in the hospital controls and hospital cases.
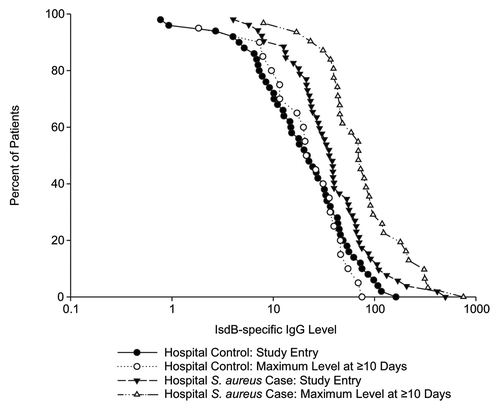
In the analysis of IsdB-specific IgG levels stratified by collection time post-infection diagnosis, the acute vs. convalescent levels were similar within a given hospital group (). Levels increased ~2.0-fold from the acute to the convalescent state in the hospital S. aureus cases. No such increase in the levels for the hospital controls was observed. Similar findings were observed when analyses were restricted to S. aureus bloodstream infections only. This is illustrated in the reverse cumulative distribution (RCD) analysis (), which shows that the acute and convalescent IgG titer distributions at baseline and at maximum differ substantially in hospital S. aureus cases, but not in hospital controls. It also illustrates that the baseline GMC for subjects with S. aureus infection were higher than for controls; this may be reflective of delayed diagnosis as no serum was obtained prior to diagnosis.
Table 4. Geometric mean fold rise in IsdB-specific IgG levels among the hospital cases and controls*.
Discussion
The clinical manifestations of S. aureus infection are extensive, ranging from uncomplicated skin and soft tissue infections to deep wound infections, pneumonia, and bacteremia. Several groups of patients have a well-defined risk for serious infections due to S. aureus; patients undergoing major surgery (including cardiothoracic and orthopedic surgery, among others) and patients undergoing chronic hemodialysis are among those populations at the highest risk.Citation22 Availability of a S. aureus vaccine for at-risk patients would represent a new paradigm in addressing this medical need, particularly in light of increasing antibiotic resistance.
IsdB is a heme-binding protein that is conserved in diverse S. aureus clinical isolates and expressed on the cell surface of the bacteria.Citation9 The potential use of this antigen as a component of a vaccine for prevention of S. aureus disease has been previously described.Citation9,Citation15 However, as a stand-alone vaccine (V710), protection was not observed in a clinical efficacy trial among cardiac surgery patients,Citation21 despite a significant increase in IsdB antibody titers in the V710 vaccinated patients. Although the IsdB specific antibody response was stimulated by V710, it remains unclear which immune mechanisms are critical to mediate clinical efficacy against S. aureus. In recently reported data, protection mediated via IsdB vaccination in mice was dependent on the T-cell (CD4+) response to the vaccine. Efficacy was mediated through an antigen specific IL17 response.Citation16 It should be noted that T-cell response to V710 (IsdB) in humans has not been evaluated.
The lack of efficacy observed in the V710 clinical trial could have as its basis two important characteristics of the target antigen, IsdB; these are antigen conservation and expression in vivo. IsdB expression is upregulated in the absence of iron, and the antigen is covalently bound to the peptidoglycan cell wall. In pre-clinical murine models, challenge with S. aureus inoculum triggers the expression of IsdB on the bacteria in the host environment within a short time post challenge.Citation9,Citation10,Citation12,Citation17 External expression on the cell wall, with exposure to elements of the host immune system, could lead to genetic variability. We have previously reported that IsdB is highly conserved among all strains examined to date.Citation9 In a recent study comparing genetic variation among 58 published genomes, McCarthy and Lindsay state that isdB is missing from at least one isolate, the USA300 TCH1516 strain.Citation23 However, when we examined the isdB, or SasJ, sequence of TCH1516, we found the sequence to be essentially identical to the isdB sequence of COL (unpublished data). Therefore, the conclusion from McCarthy and Lindsay is not supported. In our study, 51 S. aureus isolates were collected from the S. aureus cases group. These were typed and found to be from widely divergent clonal complex types (Table S1). Importantly, all isolates expressed IsdB, as demonstrated through Western staining with a murine IsdB specific mAb. A high level of conservation of the gene and antigen would be expected from this result. Additionally, expression of the antigen in the strains infecting patients in the cases group serves as the basis for the rise in IsdB-specific antibody titers observed in these patients.
Expression of a bacterial antigen, such as IsdB, during the course of natural infection among hospitalized patients is deemed to be critical to ensure its potential value as a target for a vaccine. To confirm expression in a hospitalized cohort, we examined the immune response to this antigen during confirmed infection. We utilized a well characterized immunoassay, for which performance and specificity has been described.Citation24 The assay is approximately 97% specific for antibodies to IsdB. We found that baseline anti-IsdB levels generally overlapped between the 3 groups, although the variability was fairly high within each group. Serum IgG levels to IsdB appeared to be relatively stable over an 18-mo period in the Healthy group; the correlations between baseline and follow-up levels (rho = 0.93) and between minimum and maximum levels (rho = 0.95) were strong and highly statistically significant. In the hospitalized patients, anti-IsdB levels at the last follow-up visit increased significantly from study entry levels in hospital S. aureus cases, but not in hospital controls. These data provide evidence that the IsdB protein is expressed during S. aureus infections. Importantly, the results also support the notion that patients respond immunologically to the IsdB antigen even under a state of immune depression following surgery.Citation25
It is recognized that hospitalized patients may be infected by S. aureus colonizing their nares. Colonization by S. aureus may also induce antibodies to IsdB. Although colonization data were not collected in the current study, in the Merck V710 (Protocol 003) trial, nasal colonization data was collected at day of vaccination.Citation21 Among Protocol 003 patients, it was observed that those who were nasally colonized with S. aureus, had baseline anti-IsdB titers which were somewhat higher than for patients not nasally colonized (GMC 42.2 μg/mL; 95% CI 39.8 – 44.8 μg/mL; vs. GMC 27.5 μg/mL; 95% CI 26.5–28.5 μg/mL for non- nasally colonized patients) (unpublished data). It is not known if the increase was due to colonization alone, or if the patients had previous [non-clinical] S. aureus infection, which could also increase baseline sera titers. In the current study, potential impact on patient anti-IsdB titers due to pre-existing colonization, should be reflected in the baseline titers of those patients when they were enrolled into the study. Taking this into account, a significant rise in titers was still observed during the course of disseminated infection. Collectively, the data from our study indicate that during the course of natural S. aureus infection in hospitalized patients, the bacteria express IsdB. This expression may be observed through an increase in IsdB-specific antibody titers. Moreover, all S. aureus strains examined to date express IsdB, and there is a high level of conservation of IsdB among the strains. These observations may rule out lack of target antigen as a potential explanation for the low efficacy of V710 against bacteremia and deep sternal wound infection. The inferior efficacy may be due to other unexplained factors including use of a single antigen in the vaccine, characteristics of the surgical population enrolled in the trial, or other confounding factors.
The main limitation of this analysis is that it comprised small observational studies comparing unmatched, nonrandom populations. Therefore, the ability to compare results across groups is somewhat limited. It is assumed that humans develop background titers to S. aureus infection by colonization and subclinical infection. A drawback of this study is that subjects were not monitored for nasal carriage of the organism. However, this survey was intended to be hypothesis-generating, and the significance of this finding will need to be confirmed by future studies. In addition, these results are likely to more closely resemble those that would be found in a typical, heterogeneous clinic population. Second, blood samples were drawn from symptomatic patients to confirm whether an infection was due to S. aureus. Thus, we do not know if the antibody levels at study-entry represented acute, subacute, or convalescent levels. This is further complicated by the within-group variability in entry antibody levels, as well as the overlapping ranges between groups. Third, the collection of serum samples was not standardized between the hospitalized patients, with variable number of serum samples obtained per patient at varying time points. Patients with S. aureus infection did have a slightly longer mean follow-up than patients with infections other than S. aureus (15.3 days vs. 11.6 days, p = 0.05). However, it is unlikely that the later measurements would falsely elevate the apparent rise in titer in patients with S. aureus infection since antibody titers observed in the Healthy group and patients with infections other than S. aureus were stable over time. Finally, we did not collect information on recovery or cause of death. Therefore, we cannot conclude with certainty that low antibody levels to IsdB are associated with S. aureus morbidity or mortality in humans.
In conclusion, we found that among patients with documented S. aureus infection, all isolates expressed IsdB, and that only patients with documented S. aureus infection demonstrated statistically significant increases in anti-IsdB IgG. While the anti-IsdB titers in normal healthy individuals showed no significant trend over time, the titers in patients with confirmed S. aureus infection were elevated within a few days after diagnosis. Patients with infections other than S. aureus did not have a rise in titers to IsdB as was noted in patients with S. aureus infection. Collectively, our results are supportive that IsdB was conserved and available for immunological targeting in V710 vaccinated patients.
Materials and Methods
Study populations
Sera were collected from two, independently sampled, non-matched human populations: (1) healthy adults (healthy) and (2) hospitalized patients with non-S. aureus infections (hospital controls) and patients with documented S. aureus infections (hospital S. aureus cases). This study was conducted in accordance with the principles of the Declaration of Helsinki. Informed consent was obtained for all participants, and Institutional Review Board approval was obtained for the hospitalized patients.
The healthy group included individuals who were recruited to participate in the study at two different time points (December 2003 and August 2004) to assess stability of anti-IsdB levels. These individuals were considered to be in good health and with no known incidence of S. aureus infection. Nasal colonization was not investigated. In the Healthy group, longitudinal sera (median of 3 follow-up sera, range: 1 to 3) were collected (through Merck employee Health Services, West Point, PA) with a median follow-up time of 10 mo (range: 8–18 mo) in 20 of the 21 subjects. In the hospital patient groups, eligible adult (≥ 18 y) patients, hospitalized between January 2005 and December 2006 at Queens Medical College, Nottingham, UK, who developed an infection during their hospitalization and who had not yet received antibiotic therapy were recruited to participate in the study by the investigator. Patients who had a S. aureus infection in the previous year or HIV infection/AIDS were excluded.
Hospital S. aureus cases were patients documented to have developed a S. aureus infection, and hospital controls were patients with a non-S. aureus-associated infection during their hospital stay. Sera were collected once an infection had been diagnosed but prior to the initiation of antibiotics; additional samples were taken until the patient died or was discharged from the hospital. None of the patients in the study received intravenous immunoglobulin therapy to prevent or treat a S. aureus infection. To further examine the change in IsdB-specific IgG levels during infection, levels were stratified according to collection time post diagnosis, as either acute (< 7 days post diagnosis) or convalescent (≥ 7, ≥ 10, and ≥ 14 days post diagnosis). Patients were not evaluated for nasal colonization by S. aureus.
Microbial isolates and molecular analysis
Pathogens were isolated from serum samples obtained from all of the hospital patients with documented infections, using standard blood culture techniques. Isolates were also obtained from cerebrospinal fluid, surgical tissue, synovial fluid, and sputum, using standard culture techniques. Identifications were made to the species level using standard microbiological procedures. S. aureus isolates were further speciated using spa typing as previously described.Citation26,Citation27 A spa type refers to the composition of the variable number tandem repeats in the 3′ end of the staphylococcal protein A gene (spa). Methicillin resistance was determined using standard laboratory techniques and SCCmec typing performed as previously described.Citation28
Serology assay
A direct binding assay was developed for the detection of total IgG antibodies to IsdB, using Luminex® technology.Citation24,Citation29,Citation30 The assay has been extensively characterized and used to support multiple Merck clinical trials.Citation20,Citation21,Citation31 Briefly, a truncated form of IsdB (aa 42–486), containing the two NEAT domains of IsdB (aa140–462) (750 μg) was covalently bound to 9.6 × 106 Radix maleimide-modified microspheres (Radix) via a C-terminal cysteine residue at room temperature. Microspheres were washed with phosphate buffered saline (PBS)/0.05% Tween-20 (Sigma-Aldrich) and then quenched with 1M N-acetyl-l-cysteine for 2 h at room temperature. Microspheres were washed 3× in PBS/0.05% Tween-20 and enumerated. The assay was performed in 96-well plates. Each plate contained a reference standard of pooled normal human serum containing a known anti-IsdB IgG concentration, plus three control sera. A 2-fold serial dilution of reference sera was used to generate a 12-point standard curve. Control sera, which came from individuals previously determined to have high, medium or low anti-IsdB levels, were included on each plate and diluted 1:250 and 1:2500. Antibody depleted human sera was included as a negative control. Patient serum was tested in duplicate at a 1:2500 and 1:25,000 dilution.
IsdB Expression detected by western blotting
S. aureus isolates were grown overnight in iron-restricted conditions, in Roswell Park Memorial Institute (RPMI) 1640 medium (Gibco BRL) to stationary phase. Cells were harvested by centrifugation and then lysed with a combination of lysostaphin, detergent, and sonication. Total cell lysates were loaded on 4–12% Novex Bis-Tris gradient gels (Invitrogen) and transferred to nitrocellulose. The nitrocellulose blots were probed with an IsdB specific monoclonal antibody, 2H2, previously described.Citation17 This monoclonal was demonstrated to have protective efficacy in rodent challenge models.
Statistical analysis
Since this was a descriptive study, there were no pre-defined hypotheses for comparing groups. The primary comparison of interest was the difference in geometric mean fold rises (GMFRs) between the hospital S. aureus cases and the hospital Control groups. The data from the Healthy group served mainly to provide comparative information on naturally occurring IsdB levels in non-hospitalized subjects with low risk for S. aureus infections. Descriptive statistics were used to describe the study populations and to summarize the antibody levels in the various study populations. IgG levels were transformed using natural logarithms. Differences in IgG levels from study entry to last value were estimated for each subject. The geometric mean titers concentration (GMC), and GMFR in levels and their 95% confidence intervals were estimated using the Student t-distribution. Correlation between subjects’ IgG levels was estimated using Spearman’s rank correlation coefficient (rho). Paired t-tests were used to compare mean differences in IgG levels from study entry to last value within each group. The percentage of patients with a rise in antibody to IsdB at their last visit above 2 times the standard deviation above the geometric mean of the study-entry levels for the hospital controls , a proxy for pre-infection levels, were calculated. Difference in proportions between the groups was compared using Fisher’s Exact Test.
Antibody distributions for the hospital S. aureus cases and hospital controls were described at baseline and maximum value ≥ 10 day using reverse cumulative distribution plots.
Additional material
Download Zip (101 KB)Acknowledgments
All authors are responsible for the work described in this paper, and they contributed to the conception, data interpretation, and drafting of the manuscript and/or revising the manuscript for important intellectual content. All authors provided final approval of the version to be published.
Merck Sharp and Dohme Corp., a subsidiary of Merck and Co., Inc. funded this study.
The authors thank Jennifer Pawlowski and Kim Strohmaier (Merck Sharp and Dohme Corp.) for their assistance with the preparation and submission of this manuscript. The authors also thank Jonathan Hartzel for assistance with clinical statistics from the Merck V710 Protocol 003 clinical trial.
Disclosure of Potential Conflicts of Interest
This analysis was supported by Merck and Co., Inc. JKZ, ME, MR, NK, TM, SSS, ASA, and JMA are current or former employees of Merck Sharp and Dohme and may own stock and/or stock options in the company. BNK and DDA were consultants for Merck.
Supplemental Materials
Supplemental materials may be found here: www.landesbioscience.com/journals/vaccines/article/25253
References
- Williams RE. Healthy carriage of Staphylococcus aureus: its prevalence and importance. Bacteriol Rev 1963; 27:56 - 71; PMID: 14000926
- Nouwen JL, Ott A, Kluytmans-Vandenbergh MFQ, Boelens HA, Hofman A, van Belkum A, et al. Predicting the Staphylococcus aureus nasal carrier state: derivation and validation of a “culture rule”. Clin Infect Dis 2004; 39:806 - 11; http://dx.doi.org/10.1086/423376; PMID: 15472812
- Nilsson P, Ripa T. Staphylococcus aureus throat colonization is more frequent than colonization in the anterior nares. J Clin Microbiol 2006; 44:3334 - 9; http://dx.doi.org/10.1128/JCM.00880-06; PMID: 16954269
- Steinberg JP, Clark CC, Hackman BO. Nosocomial and community-acquired Staphylococcus aureus bacteremias from 1980 to 1993: impact of intravascular devices and methicillin resistance. Clin Infect Dis 1996; 23:255 - 9; http://dx.doi.org/10.1093/clinids/23.2.255; PMID: 8842259
- Rihn JA, Michaels MG, Harner CD. Community-acquired methicillin-resistant staphylococcus aureus: an emerging problem in the athletic population. Am J Sports Med 2005; 33:1924 - 9; http://dx.doi.org/10.1177/0363546505283273; PMID: 16314668
- Shukla SK. Community-associated methicillin-resistant Staphylococcus aureus and its emerging virulence. Clin Med Res 2005; 3:57 - 60; http://dx.doi.org/10.3121/cmr.3.2.57; PMID: 16012120
- Lowy FD. Staphylococcus aureus infections. N Engl J Med 1998; 339:520 - 32; http://dx.doi.org/10.1056/NEJM199808203390806; PMID: 9709046
- Mazmanian SK, Skaar EP, Gaspar AH, Humayun M, Gornicki P, Jelenska J, et al. Passage of heme-iron across the envelope of Staphylococcus aureus. Science 2003; 299:906 - 9; http://dx.doi.org/10.1126/science.1081147; PMID: 12574635
- Kuklin NA, Clark DJ, Secore S, Cook J, Cope LD, McNeely T, et al. A novel Staphylococcus aureus vaccine: iron surface determinant B induces rapid antibody responses in rhesus macaques and specific increased survival in a murine S. aureus sepsis model. Infect Immun 2006; 74:2215 - 23; http://dx.doi.org/10.1128/IAI.74.4.2215-2223.2006; PMID: 16552052
- Pancari G, Fan H, Smith S, Joshi A, Haimbach R, Clark D, et al. Characterization of the mechanism of protection mediated by CS-D7, a monoclonal antibody to Staphylococcus aureus iron regulated surface determinant B (IsdB). Front Cell Infect Microbiol 2012; 2:36; http://dx.doi.org/10.3389/fcimb.2012.00036; PMID: 22919628
- Skaar EP, Schneewind O. Iron-regulated surface determinants (Isd) of Staphylococcus aureus: stealing iron from heme. Microbes Infect 2004; 6:390 - 7; http://dx.doi.org/10.1016/j.micinf.2003.12.008; PMID: 15101396
- Pishchany G, Dickey SE, Skaar EP. Subcellular localization of the Staphylococcus aureus heme iron transport components IsdA and IsdB. Infect Immun 2009; 77:2624 - 34; http://dx.doi.org/10.1128/IAI.01531-08; PMID: 19398548
- Ebert T, Smith S, Pancari G, Clark D, Hampton R, Secore S, et al. A fully human monoclonal antibody to Staphylococcus aureus iron regulated surface determinant B (IsdB) with functional activity in vitro and in vivo. Hum Antibodies 2010; 19:113 - 28; PMID: 21178283
- Ebert T, Smith S, Pancari G, Wu X, Zorman J, Clark D, et al. Development of a rat central venous catheter model for evaluation of vaccines to prevent Staphylococcus epidermidis and Staphylococcus aureus early biofilms. Hum Vaccin 2011; 7:630 - 8; http://dx.doi.org/10.4161/hv.7.6.15407; PMID: 21508677
- Stranger-Jones YK, Bae T, Schneewind O. Vaccine assembly from surface proteins of Staphylococcus aureus. Proc Natl Acad Sci U S A 2006; 103:16942 - 7; http://dx.doi.org/10.1073/pnas.0606863103; PMID: 17075065
- Joshi A, Pancari G, Cope L, Bowman EP, Cua D, Proctor RA, et al. Immunization with Staphylococcus aureus iron regulated surface determinant B (IsdB) confers protection via Th17/IL17 pathway in a murine sepsis model. Hum Vaccin Immunother 2012; 8:336 - 46; http://dx.doi.org/10.4161/hv.18946; PMID: 22327491
- Brown M, Kowalski R, Zorman J, Wang XM, Towne V, Zhao Q, et al. Selection and characterization of murine monoclonal antibodies to Staphylococcus aureus iron-regulated surface determinant B with functional activity in vitro and in vivo. Clin Vaccine Immunol 2009; 16:1095 - 104; http://dx.doi.org/10.1128/CVI.00085-09; PMID: 19553551
- Kim HK, DeDent A, Cheng AG, McAdow M, Bagnoli F, Missiakas DM, et al. IsdA and IsdB antibodies protect mice against Staphylococcus aureus abscess formation and lethal challenge. Vaccine 2010; 28:6382 - 92; http://dx.doi.org/10.1016/j.vaccine.2010.02.097; PMID: 20226248
- Cheng AG, Kim HK, Burts ML, Krausz T, Schneewind O, Missiakas DM. Genetic requirements for Staphylococcus aureus abscess formation and persistence in host tissues. FASEB J 2009; 23:3393 - 404; http://dx.doi.org/10.1096/fj.09-135467; PMID: 19525403
- Harro CD, Betts RF, Hartzel JS, Onorato MT, Lipka J, Smugar SS, et al. The immunogenicity and safety of different formulations of a novel Staphylococcus aureus vaccine (V710): results of two Phase I studies. Vaccine 2012; 30:1729 - 36; http://dx.doi.org/10.1016/j.vaccine.2011.12.045; PMID: 22192849
- Fowler VG, Allen KB, Moreira ED, Moustafa M, Isgro F, Boucher HW, et al. Effect of an investigational vaccine for preventing Staphylococcus aureus infections after cardiothoracic surgery: a randomized trial. JAMA 2013; 309:1368 - 78; http://dx.doi.org/10.1001/jama.2013.3010; PMID: 23549582
- Kessler M, Hoen B, Mayeux D, Hestin D, Fontenaille C. Bacteremia in patients on chronic hemodialysis. A multicenter prospective survey. Nephron 1993; 64:95 - 100; http://dx.doi.org/10.1159/000187285; PMID: 8502343
- McCarthy AJ, Lindsay JA. Genetic variation in Staphylococcus aureus surface and immune evasion genes is lineage associated: implications for vaccine design and host-pathogen interactions. BMC Microbiol 2010; 10:173; http://dx.doi.org/10.1186/1471-2180-10-173; PMID: 20550675
- Raedler MD, Heyne S, Wagner E, Shalkowski SK, Secore S, Anderson AS, et al. Serologic assay to quantify human immunoglobulin G antibodies to the Staphylococcus aureus iron surface determinant B antigen. Clin Vaccine Immunol 2009; 16:739 - 48; http://dx.doi.org/10.1128/CVI.00478-08; PMID: 19321693
- Mekontso-Dessap A, Honoré S, Kirsch M, Plonquet A, Fernandez E, Touqui L, et al. Blood neutrophil bactericidal activity against methicillin-resistant and methicillin-sensitive Staphylococcus aureus during cardiac surgery. Shock 2005; 24:109 - 13; http://dx.doi.org/10.1097/01.shk.0000171871.50524.81; PMID: 16044079
- Shopsin B, Gomez M, Montgomery SO, Smith DH, Waddington M, Dodge DE, et al. Evaluation of protein A gene polymorphic region DNA sequencing for typing of Staphylococcus aureus strains. J Clin Microbiol 1999; 37:3556 - 63; PMID: 10523551
- Shopsin B, Gomez M, Waddington M, Riehman M, Kreiswirth BN. Use of coagulase gene (coa) repeat region nucleotide sequences for typing of methicillin-resistant Staphylococcus aureus strains. J Clin Microbiol 2000; 38:3453 - 6; PMID: 10970402
- Chen L, Mediavilla JR, Oliveira DC, Willey BM, de Lencastre H, Kreiswirth BN. Multiplex real-time PCR for rapid Staphylococcal cassette chromosome mec typing. J Clin Microbiol 2009; 47:3692 - 706; http://dx.doi.org/10.1128/JCM.00766-09; PMID: 19726600
- Jones LP, Zheng HQ, Karron RA, Peret TCT, Tsou C, Anderson LJ. Multiplex assay for detection of strain-specific antibodies against the two variable regions of the G protein of respiratory syncytial virus. Clin Diagn Lab Immunol 2002; 9:633 - 8; PMID: 11986272
- Waterboer T, Sehr P, Michael KM, Franceschi S, Nieland JD, Joos TO, et al. Multiplex human papillomavirus serology based on in situ-purified glutathione s-transferase fusion proteins. Clin Chem 2005; 51:1845 - 53; http://dx.doi.org/10.1373/clinchem.2005.052381; PMID: 16099939
- Harro C, Betts R, Orenstein W, et al. A Phase I, Dose-Ranging Study to Evaluate Immunogenicity and Safety of a Novel Staphylococcus aureus Vaccine (V710). Abstracts of the 13th International Symposium on Staphylococci and Staphylococcal Infections 2008.