Abstract
Host genome is still poorly investigated in the context of vaccine or immunotherapy, however recently findings emphasized that it may affect the response to those treatments. In our retrospective study we evaluated the effect of HIV-1 genetic restriction factors on the response to dendritic cell (DC)-based immunotherapy in a Brazilian cohort of HIV positive (HIV+) patients that underwent a phase I clinical trial in 2004.
Genomic DNA from 18 HIV+ individuals that underwent DC-based immunotherapy was analyzed for selected polymorphisms known to be associated with susceptibility to HIV-1 infection and/or AIDS progression. Allelic and genotypic distribution of the 22 polymorphisms was evaluated considering the response to the treatment.
The rs11884476 SNP in PARD3B resulted associated with good response to immune treatment according to an over-dominant model. Even if functional effect of this variation is still unknown, our data suggested that it could play a role in the control of viral replication.
Our findings, being aware of the limitation represented by the small number of subjects analyzed, suggest that genetic factors involved in AIDS progression could affect the response to immunotherapy, reinforcing the idea that deeper investigation on host genetic variations will be fundamental for a rational vaccine development.
Dendritic cell (DC)-based immune treatments have gained great interest as alternative therapy for HIV-1 infected (HIV+) individuals as they are conceived to induce durable cellular responses to control viral replication through an autologous, safe and well-tolerated protocol. However, at present, only few clinical trials reported a significant control of plasma viral load (PVL) in HIV+ patients.Citation1,Citation2
In 2004 Lu et al.Citation3 performed a phase I clinical trial of DC-based treatment on 18 Brazilian HIV+ patients. Three doses of autologous monocyte-derived DC pulsed with autologous chemically inactivated HIV-1 were administered every 15 d and PVL were monitored up to 1 y. Eight out of 18 individuals showed prolonged suppression of PVL (Good Responders, GR; >90% PVL decrease 1 y after immunization), whereas 10 did not (Weak or Transient Responders, WTR; <90% PVL decrease 1 y after immunization), opening discussion about factors generating this differential response.
Host genetic background has been reported to affect individual response to prophylactic anti-HIV vaccinesCitation4,Citation5 emphasizing that vaccine-induced cellular immunity and natural immune control against the virus share common genetic contributors. Similarly, our group showed that single nucleotide polymorphisms (SNPs) in innate immune genes, specifically MBL2 and NOS1, were associated with individual response to DC-based immune treatment of Lu et al.Citation6
Another point of interest concerns the genotyping of known host anti-HIV restriction factors in patients enrolled in DC-based clinical trials because of the natural impact on viral replication in these individuals. We hypothesized that individuals with a less permissive genomic profile for HIV-1 infection and/or replication may respond better to DC-based treatment.
To evaluate the impact of host anti-HIV restriction factors on the efficacy of DC-based treatment, we analyzed the 18 HIV+ patients that participate in Lu clinical trialCitation3 for selected polymorphisms in genes known to be involved in HIV-1 infection and/or progression to AIDS.
Genomic DNA of those patients was already available and its quality/quantity checked at our laboratory using both spectrophotometer and Nanodrop. Full clinical data of the patients are available in Lu et al.,Citation3 whereas characteristic considered relevant for this study were summarized in Table S1.
Twenty two polymorphisms in 13 genes involved in HIV-1 host restriction (APOBEC3G, CCL4, CCL5, CCR5, CUL5, CXCR6, HLA-C, IFNG, PARD3B, Prox1, SDF-1, TRIM5, ZNRD1) were selected based on previously published data (briefly revised in ref. Citation7; reported in Table S2). Genotyping was performed using commercially available TaqMan assays (Applied Biosystems/AB) and ABI7500 Real-Time platform (AB). Allelic discrimination was performed using the SDS v1.4 Software (AB). CCR5 ∆32 deletion was evaluated by PCR-RFLP. PARD3B genotyping results have been double checked and confirmed by direct sequencing of the amplicon containing the rs11884476 SNP.
The frequency of PARD3B rs11884476 SNP was then evaluated in a population coming from Recife (same metropolitan area of the Brazilian clinical trialCitation3). 119 HIV+ patients (34 males, average age = 40.4; 85 females, average age = 34.95) and 212 healthy controls (HC; 59 males, average age = 24.91; 153 females, average age = 32.04) were recruited respectively at the Immunologic Day Hospital of the “Instituto de Medicina Integral Prof. Fernando Figueira” (IMIP), and at the Transfusion Center HEMOPE of Recife.
R-project software was used to calculate allelic, genotypic and haplotypic frequencies, P values and Odds Ratio (OR) as well as for genotypes modeling (package “SNP assoc” version 1.5–2). Haploview software was employed to derivate haplotypes. Polymorphisms frequencies were compared with Chi-square test with Yate’s continuity correction, which accounts for adjusting the P values of comparisons between data sets with a small number of observations in a genotype class (even less than five). Comparison between PARD3B rs11884476 genotypes and immunologic characteristic of the 18 patients submitted to immune treatment (found in Citation3), such as PVL, CD4+ and CD8+ cell counts as well as IFN-γ positive cells, was done by t test using GraphPad Prism software. Genevar software was used to evaluate in silico the functional impact of PARD3B rs11884476 on mRNA biology.
The selected 22 polymorphisms in 13 HIV-1 restriction factor genes were genotyped in the 18 HIV+ individuals submitted to the Brazilian phase I clinical trial of DC-based treatment.Citation3 Allelic and genotypic frequencies were in Hardy-Weinberg equilibrium. Polymorphisms distribution was compared in good responder and weak or transient responder according to the classification applied by Lu et al.Citation3 ().
Table 1. Results of polymorphisms association analysis in 18 HIV+ patients underwent DC-based immune treatment against HIV-1 classified in good responder (GR) and weak or transient responder (WTR) according to Lu et al.Citation3
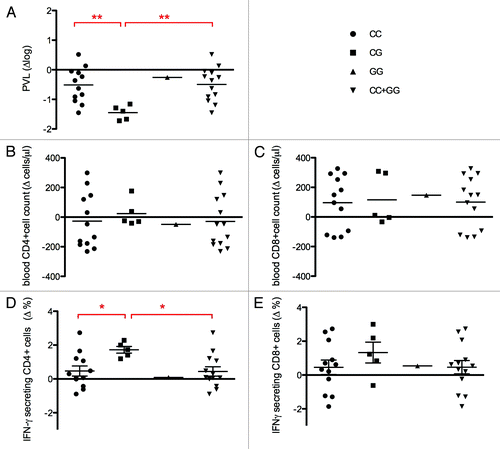
The rs11884476 polymorphism in PARD3B resulted associated with good response to the immune treatment according to an over-dominant model (C/G vs. C/C+G/G; P = 6.5exp-3), being C/G more frequent in GR than in WTR (5/8 vs. 0/10).
When changes in PVL and cellular response were stratified according to patients’ PARD3B genotypes (Table S1), rs11884476 C/G genotype was associated with greater PVL reduction compared with C/C or C/C+G/G (P = 4.3exp-3 or P = 2.6exp-3). Similarly, the rs11884476 C/G was associated with higher increase in IFN-γ producing CD4+ cells compared with C/C (P = 0.021) or C/C+G/G (P = 0.015) (). These findings are in agreement with previously reported data about the association of rs11884476 with better prognostic and delayed AIDS.Citation8
Figure 1. Plasma viral load (PVL) reduction and cellular response in 18 HIV+ patients who’s underwent DC-based immune treatment against HIV-1 according to PARD3B rs11884476 genotypes. Change in PVL expressed as log change (∆log), change in CD4+ and CD8+ cells counts (∆cells/µl) and change in percentage of CD4+ and CD8+ cells producing IFN-ϒ (∆%) are reported for the 18 HIV+ patients included in the phase I clinical trial of DC-based immune-therapyCitation3 classified according to PARD3B rs11884476 genotypes. The data, obtained from Lu et al.,Citation3 represent difference (∆) between values presented 1 y after immunization and before the starting of the trial. Individual data and media were reported. (A) Plasma viral load (PVL). Individual blood CD4+ (B) and CD8+ (C) cell counts. (D and E) Intracellular IFN-γ detection of T cells following stimulation with HIV-1-pulsed DC. Percentage of total CD4+ (D) or CD8+ (E) cell secreting IFN-γ is reported. T test analysis was performed between C/C and C/G groups and between C/C+G/G and C/G groups according to an over-dominant model. Being unique value the G/G has been excluded from the analysis. *P < 0.05; **P < 0.01.
The rs11884476 SNP is an intronic variant, with still unknown functional effect, even if it could be a tag for other polymorphisms such as for the rs10185378 reported to be associated to an alternative mRNA splicing of PARD3B gene.Citation8 As biologic samples, other than genomic DNA, from those 18 patients were no longer available and functional studies were not possible, we evaluated the possible impact of the SNP on PARD3B mRNA levels in silico using the GENEVAR database (http://www.sanger.ac.uk/humgen/genevar/), able to display mRNA expression profiles of B lymphoblastoid cell lines established from HapMap donors. The unique data available for rs11884476 genotypes are referring to YRI population and they did not significantly correlate with PARD3B mRNA level variation (P = 0.181) (Fig. S3), suggesting that, in this population, this SNP could not affect mRNA production.
To investigate the prevalence of this polymorphism in HIV+ population from the same geographical region of the 18 patients submitted to immune treatment,Citation3 PARD3B rs11884476 was then genotyped in a case/control study (n = 119/212) with patients and controls coming from Recife. We did not find any significant association between PARD3B rs11884476 SNP and susceptibility to HIV-1 infection (Table S4), however it is interesting to emphasize that the HIV+ cohort enrolled for the study were not efficient controllers of virus replication, being PVL always greater than 2 log copy/ml before starting the HAART treatment, whereas PARD3B rs11884476 was previously associated with delayed AIDS.Citation8
Allelic and genotypic frequencies of the 18 patients submitted to DC immune-treatment (classified as GR and WTR) were then compared with those of HIV+ patients form Recife. No significant difference was found between WTR and HIV+ patients, while a significant difference has been observed between GR and HIV+ patients considering the over-dominant model (P = 0.025) (Table S4), reinforcing the idea that individuals with rs11884476 in heterozygosis were able to control viral replication more efficiently than individuals homozygotes for the SNP, and that these “controllers” of virus replication may have a greater chance to better respond to immune treatment.
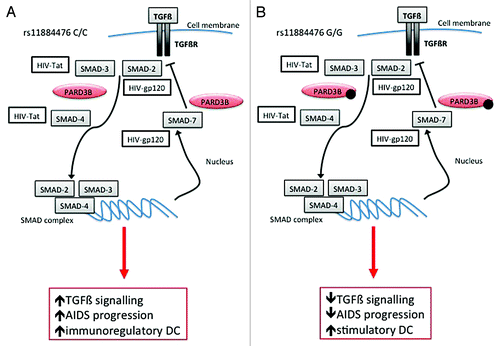
PARD3B protein interacts with TGFß signaling proteins SMAD, which directly binds HIV-1 proteins Tat and gp120.Citation8 As increasing levels of TGFß are typically detected during HIV-1 replication and progression to AIDS, rs11884476 variant could affect PARD3B-SMAD interaction resulting in TGFß signaling down regulation, leading to better control of AIDS progression. However, reducing production of TGFß is recommended in HIV vaccine design due to its immunomodulatory function on DC activation,Citation9 suggesting that polymorphisms in PARD3B could affect both AIDS progression as well as DC-mediated lymphocytes activation.
The possible dual role of PARD3B in terms of viral replication and AIDS progression, or DC-mediated lymphocytes activation is depicted in the cartoon reported in . According to the over-dominant model, we may hypothesize that individual heterozygotes for rs11884476 SNP could have a TGFß signaling leading to a balance between regulatory and stimulatory DC profile.
Figure 2. Possible interactions between PARD3B and SMAD proteins influencing TGFß signaling. The two hypotheses concerning the dual role of PARD3 in terms of viral replication control or DC-mediated lymphocytes activation are reported according to rs11884476 genotypes (2A: wild type C/C genotype; 2B: G/G genotype). The up or downregulation of TGFß signaling and the consequences in terms of AIDS progression or DC immune-regulation are evidenced in red rectangles.
Despite some limitations of this study, such as the very low number of individuals analyzed for few genetic variants and the lack of biologic samples to deeper corroborate our data, our findings lead us to hypothesize that genetic factors involved in AIDS progression could affect the response to therapeutic DC vaccine, reinforcing the idea that deeper investigation on host genetic variations will be fundamental for a rational vaccine development.
Additional material
Download Zip (153.6 KB)Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
This work was supported by the Pernambuco Research Foundation (FACEPE), Sao Paulo Research Foundation (FAPESP, n. 2013/06142-1) and Brazilian National Council for Scientific and Technologic Development (CNPq).
References
- García F, Routy JP. Challenges in dendritic cells-based therapeutic vaccination in HIV-1 infection Workshop in dendritic cell-based vaccine clinical trials in HIV-1. Vaccine 2011; 29:6454 - 63; http://dx.doi.org/10.1016/j.vaccine.2011.07.043; PMID: 21791232
- García F, Climent N, Guardo AC, Gil C, León A, Autran B, Lifson JD, Martínez-Picado J, Dalmau J, Clotet B, et al, DCV2/MANON07-ORVACS Study Group. A dendritic cell-based vaccine elicits T cell responses associated with control of HIV-1 replication. Sci Transl Med 2013; 5:ra2; http://dx.doi.org/10.1126/scitranslmed.3004682; PMID: 23283367
- Lu W, Arraes LC, Ferreira WT, Andrieu JM. Therapeutic dendritic-cell vaccine for chronic HIV-1 infection. Nat Med 2004; 10:1359 - 65; http://dx.doi.org/10.1038/nm1147; PMID: 15568033
- Kulkarni H, Marconi VC, Agan BK, McArthur C, Crawford G, Clark RA, Dolan MJ, Ahuja SK. Role of CCL3L1-CCR5 genotypes in the epidemic spread of HIV-1 and evaluation of vaccine efficacy. PLoS One 2008; 3:e3671; http://dx.doi.org/10.1371/journal.pone.0003671; PMID: 18989363
- Fellay J, Frahm N, Shianna KV, Cirulli ET, Casimiro DR, Robertson MN, Haynes BF, Geraghty DE, McElrath MJ, Goldstein DB, National Institute of Allergy and Infectious Diseases Center for HIV/AIDS Vaccine Immunology, NIAID HIV Vaccine Trials Network. Host genetic determinants of T cell responses to the MRKAd5 HIV-1 gag/pol/nef vaccine in the step trial. J Infect Dis 2011; 203:773 - 9; http://dx.doi.org/10.1093/infdis/jiq125; PMID: 21278214
- Segat L, Brandão LA, Guimarães RL, Pontillo A, Athanasakis E, de Oliveira RM, Arraes LC, de Lima Filho JL, Crovella S. Polymorphisms in innate immunity genes and patients response to dendritic cell-based HIV immuno-treatment. Vaccine 2010; 28:2201 - 6; http://dx.doi.org/10.1016/j.vaccine.2009.12.056; PMID: 20056178
- An P, Winkler CA. Host genes associated with HIV/AIDS: advances in gene discovery. Trends Genet 2010; 26:119 - 31; http://dx.doi.org/10.1016/j.tig.2010.01.002; PMID: 20149939
- Troyer JL, Nelson GW, Lautenberger JA, Chinn L, McIntosh C, Johnson RC, Sezgin E, Kessing B, Malasky M, Hendrickson SL, et al. Genome-wide association study implicates PARD3B-based AIDS restriction. J Infect Dis 2011; 203:1491 - 502; http://dx.doi.org/10.1093/infdis/jir046; PMID: 21502085
- Kornbluth RS, Stone GW. Immunostimulatory combinations: designing the next generation of vaccine adjuvants. J Leukoc Biol 2006; 80:1084 - 102; http://dx.doi.org/10.1189/jlb.0306147; PMID: 16931603
