Abstract
Development of vaccines for preventing a future pandemic of severe acute respiratory syndrome (SARS) caused by SARS coronavirus (SARS-CoV) and for biodefense preparedness is urgently needed. Our previous studies have shown that a candidate SARS vaccine antigen consisting of the receptor-binding domain (RBD) of SARS-CoV spike protein can induce potent neutralizing antibody responses and protection against SARS-CoV challenge in vaccinated animals. To optimize expression conditions for scale-up production of the RBD vaccine candidate, we hypothesized that this could be potentially achieved by removing glycosylation sites in the RBD protein. In this study, we constructed two RBD protein variants: 1) RBD193-WT (193-aa, residues 318–510) and its deglycosylated forms (RBD193-N1, RBD193-N2, RBD193-N3); 2) RBD219-WT (219-aa, residues 318–536) and its deglycosylated forms (RBD219-N1, RBD219-N2, and RBD219-N3). All constructs were expressed as recombinant proteins in yeast. The purified recombinant proteins of these constructs were compared for their antigenicity, functionality and immunogenicity in mice using alum as the adjuvant. We found that RBD219-N1 exhibited high expression yield, and maintained its antigenicity and functionality. More importantly, RBD219-N1 induced significantly stronger RBD-specific antibody responses and a higher level of neutralizing antibodies in immunized mice than RBD193-WT, RBD193-N1, RBD193-N3, or RBD219-WT. These results suggest that RBD219-N1 could be selected as an optimal SARS vaccine candidate for further development.
Introduction
Severe acute respiratory syndrome (SARS) was first diagnosed in Guangdong Province, China, in 2002, ultimately spreading to 29 countries where it caused 776 deaths and over 8000 respiratory infections.Citation1 Researchers identified the SARS coronavirus (SARS-CoV) as the etiologic agent of SARS in 2003Citation2,Citation3 and subsequently the National Institute of Allergy and Infectious Diseases (NIAID) of the US. National Institutes of Health (NIH) labelled it a Category C pathogen, along with other highly transmissible agents of potential biodefense importance.Citation4
Because of the explosive nature of the 2002–03 SARS pandemic, an intensive effort has been underway to develop SARS countermeasures, including vaccines.Citation1 A stable and effective SARS-CoV vaccine could be stockpiled as part of national or global public health emergency preparedness efforts.Citation4 Initial efforts focused on developing whole virus vaccines that were often inactivated by chemical agents or radiation and adjuvanted on alumCitation1 However, in laboratory mice, it was observed that such vaccines elicited eosinophilic immunoenhancing pathology with evidence of TH2-linked alveolar damage.Citation5,Citation6 Previously, immune enhancing pathology in vaccinated children derailed similar efforts to develop inactivated respiratory syncytial virus (RSV) vaccines.Citation7
As an alternative approach, prototype subunit vaccines comprised of the SARS-CoV spike (S) protein have been developed.Citation1 Like HIV gp160 and influenza hemagglutinin, the SARS-CoV S protein is a class I viral fusion protein, and, as such, it is a major target of host neutralizing antibodies.Citation1,Citation4 Efforts to develop genetically engineered SARS-CoV S protein vaccines were reviewed previously.Citation1 Briefly, both baculovirus-expressed recombinant protein adjuvanted with alum and a Venezuelan equine encephalitis vector containing S-protein plasmid were shown to elicit protection in BALB/c mice challenged with live SARS-CoV,Citation1,Citation8 but some S-protein constructs expressed in mammalian cells were found to cause antibody-mediated enhancement.Citation9
As a substitute for the full-length S protein, its 193 amino acids (aa) minimal receptor-binding domain containing residues 318–510 (RBD193) was identified and found to bind to its putative human receptor, a transmembrane angiotensin-converting enzyme 2 (ACE2), in vitro.Citation10 In addition, recombinant proteins RBD193 and a related construct, RBD219 (residues 318–536), expressed in the culture supernatant of mammalian cells 293T and Chinese hamster ovary (CHO)-K1, respectively, were demonstrated to elicit neutralizing antibodies and protective immunity in vaccinated mice.Citation11,Citation12 Moreover, RBD can also absorb neutralizing antibodies in the antisera of mice, monkeys, and rabbits immunized with whole SARS-CoV or vaccinia virus expressing S protein constructs.Citation13
Here we report the expression of RBD193 and RBD219 recombinant proteins, as well as their deglycosylated forms in the yeast Pichia pastoris. We found that one of the proteins, RBD219-N1, in which an N-linked glycosylated asparagine at the N-1 position of RBD219 had been deleted, could be expressed and purified in high yield, and maintained its functionality and antigenicity as the mammalian cell-expressed RBD193. RBD219-N1 elicited high titers of neutralizing antibodies against both SARS-CoV pseudovirus and live virus. Therefore, this molecule has been selected for scale-up process development and manufacture as a recombinant SARS vaccine for clinical testing.
Results
Expression of recombinant RBDs in P. pastoris
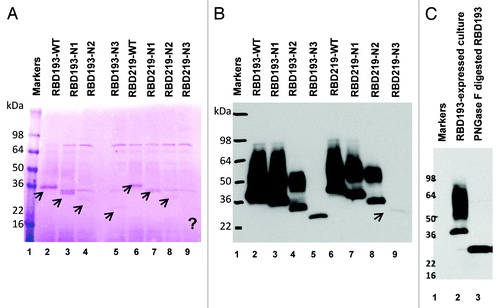
Different constructs of RBD193 and RBD219 (WT, N1, N2, and N3) () were transformed into P. pastoris X-33, and 20 clones from each transformation were induced for recombinant protein expression in 10 ml tubes with 0.5% methanol. After induction for 72 h, the constructed recombinant RBDs with different size were observed by SDS-PAGE Coomassie-stained gels and western blot with anti-RBD monoclonal antibody (mAb) 33G4 previously produced in our laboratory.Citation14 The apparent molecular weight (M.W.) of recombinant RBDs was higher than expected based on the sequence, and a high M.W. smear was observed, especially in wild-type (WT) constructs, in both RBD193 and RBD219, indicating that the recombinant RBD-WTs were glycosylated or aggregated (). The extent of glycosylation of RBD193-WT was confirmed by digesting the protein with N-glycosidase PNGase F. After digestion, the high M.W. smear disappeared, and the size of RBD193-WT returned to the expected M.W. (23 kDa) (). This assay also confirmed that the high M.W. smear was from high glycosylation of the yeast-expressed RBDs, and not from aggregation. Further evidence for the glycosylation of yeast-expressed RBDs was confirmed when the N-linked glycosylation sites were deleted or mutated (N1, N2, and N3); both the extent of glycosylation and the apparent M.W. of recombinant RBDs were reduced (), accordingly. These constructs with the deglycosylated forms allowed accurate and reproducible control during scale-up production and quality control testing. Notably, successively decreased expression yield of recombinant RBDs was also observed as N-linked glycosylation sites were deleted/mutated (yield level: WT > N1 > N2 > N3; ). This suggested that a workable balance between expression yield and glycosylation level should be considered during product development.
Figure 1. Schematic diagram of the different SAR-CoV S-RBD protein expression constructs. Both RBD193-WT and RBD219-WT contain 3 N-glycosylation sites, including N-1, N-13, and N-40. The deleted or mutated N-glycosylation sites are highlighted with a line crossing the amino acid (e.g., N-1) or italics (e.g., S-13, and A-40), respectively.

Figure 2. Expression profiles of the different SAR-CoV RBD protein constructs in yeast. The expression of wild-type (WT) and the different deglycosylated proteins of RBD193 and RBD219 in P. pastoris X-33 after induction with methanol were detected by SDS-PAGE (A) and by western blot with anti-RBD mAb 33G4 (0.2 µg/ml) (B). Each lane was loaded with 10 µl of induced culture (unpurified). (C) The N-linked glycan on yeast-expressed recombinant RBD193-WT could be removed completely by Peptide-N-Glycosidase F (PNGase F) digestion. Lane 1: protein marker, lane 2: RBD193-expressed culture (10 µl), lane 3: PNGase F digested RBD193.
Optimization of expression condition: pH and detergent
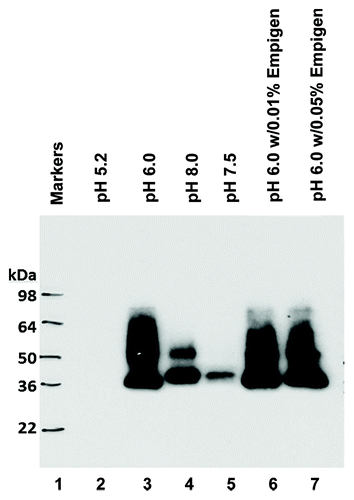
To maximize the expression yield and minimize the possible aggregation of recombinant RBDs in P. pastoris X33, we used RBD193-WT as the prototype to test the optimized induction conditions using media with different pH and/or different concentration of Empigen detergent. Based on western blot with anti-RBD mAb 33G4, the optimal pH for RBD193 expression was pH 6.0. No target protein was expressed in culture with pH 5.0, and even less expression of RBD193 was seen in media with pH over 6.0. The addition of (0.01% or 0.05%) Empigen detergent did not improve the expression yield or change the pattern of expressed RBD, indicating that aggregation does not affect expressed RBD193-WT ().
Figure 3. Detection of the expression of RBD193-WT protein in yeast by western blot. RBD193-WT was induced in P. pastoris culture with different pH (Lane 1: markers; Lane 2: pH 5.2; Lane 3: pH 6.0; Lane 4: pH 7.5; and Lane 5: pH 8.0) and with different amounts of detergent (Lane 6: pH 6.0 w/0.01% Empigen and lane 7: pH 6.0 w/0.05% Empigen). The expressed proteins in the medium were transferred on PVDF and probed with 0.2 µg/ml of anti-RBD mAb 33G4.
Fermentation and purification of RBD constructs
The RBD193-WT yeast construct stopped growing when being induced with methanol for 48 h, as a possible result of the toxicity of expressed RBD193-WT to the yeast cells. Considering the low expression yield of RBD193-N2, RBD219-N2, and RBD219-N3, we excluded them for further scaling-up study. Consequently, four other constructs, including different deglycosylated forms (RBD193-N1, RBD193-N3, RBD219-WT, and RBD219-N1), were chosen for the 5 L scale fermentation in order to obtain recombinant proteins for comparison of immunogenicity and potency. After 5 L fermentation, recombinant RBD193-N1, RBD219-WT, and RBD219-N1 were purified from the fermented culture by Butyl HP and size exclusion chromatography (SEC), and recombinant RBD193-N3 was purified by negative capture on Q Sepharose XL followed by SEC.
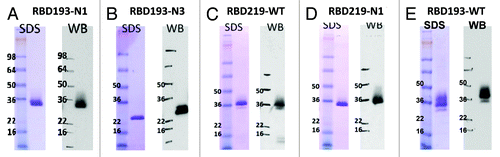
Initial attempts to purify RBD193-N1, RBD219-WT, and RBD219-N1 from the fermented culture using hydrophobic interaction (Butyl HP) chromatography and subsequent size exclusion column resulted with 95% of purified proteins, as evidenced by both SDS-PAGE Coomassie-stained gels and western blot with anti-RBD mAb 33G4 (). Moreover the highly glycosylated RBD and host protein contaminants could be efficiently removed with this two-step purification procedure, resulting with reliably pure and soluble products. Unlike the other expressed RBD proteins using other expression systems,Citation11,Citation12,Citation15 these yeast-expressed RBDs do not contain a 6 × His tag or any other fusion tag, thus permitting future scale-up manufacture for human use.
Figure 4. SDS-PAGE and western blot analysis of yeast-expressed RBD proteins. SDS-PAGE (SDS, left panels) and western blot (WB, right panels) analysis of 2 μg purified RBD 193-N1 (A), RBD193-N3 (B), RBD219-WT (C), and RBD219-N1 (D) were performed. Western blot was probed with 0.2 µg/ml of anti-RBD mAb 33G4. Wild-type SARS-CoV RBD protein expressed in 293T cells (RBD193-WT) was included as the positive control (E).
Antigenicity analysis of yeast-expressed RBDs of SARS-CoV
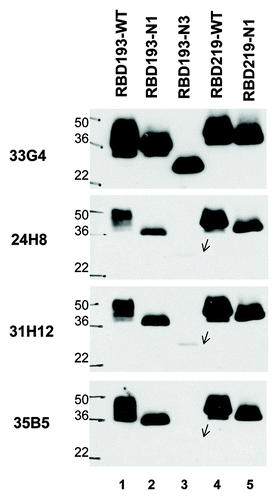
In order to confirm whether the selected purified RBD constructs (RBD193-N1, RBD193-N3, RBD219-WT, and RBD219-N1) shared the same antigenicity or antigenic epitopes with the mammalian cell (293T) expressed-RBD that we know can elicit the production of neutralizing antibody,Citation11,Citation12 we performed western blot against four conformational anti-RBD mAbs, including 24H8 (Conf I), 31H12 (Conf II), 35B5 (Conf IV), and 33G4 (Conf V)Citation14 (). Notably, mAb 33G4 strongly recognized all the RBD constructs, similarly to the recognition pattern with the mammalian cell-expressed RBD, while mAb 24H8 recognized these proteins only weakly. Furthermore, all yeast-expressed RBD219 constructs could be recognized more strongly by the four specific mAbs when the film exposure time was increased (data not shown), in contrast to the RBD193 constructsCitation11,Citation12 and in agreement with previous results. This line of evidence strongly suggests that the wild-type and deglycosylated constructs exhibit similar antigenicity.
Figure 5. Antigenicity of yeast-expressed RBD proteins. To detect antigenicity of yeast-expressed RBDs (2 µg for each protein), we used mAbs specific for the conformational epitopes of SARS-CoV RBD and western blot. The mAbs 35B5 (Conf IV), 33G4 (Conf V), 24H8 (Conf I), and 31H12 (Conf II) at 0.2 µg/ml were used for the test. Wild-type SARS-CoV RBD protein expressed in 293T cells (RBD193-WT) was included as the positive control. Protein molecular weight marker (Marker) was indicated on the left.
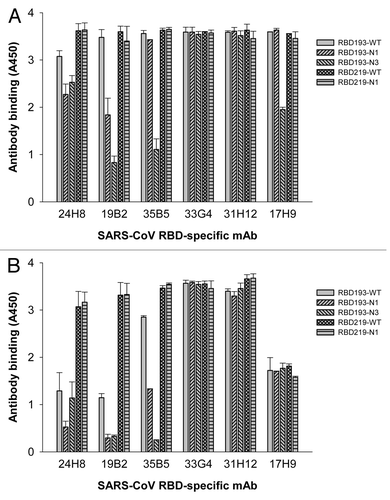
To further validate the antigenicity of these RBD proteins, we performed ELISA with five conformational anti-RBD mAbs (24H8, 31H12, 35B5, 33G4, and 19B2) and one linear anti-RBD mAb (17H9).Citation14 As shown in , while all yeast-expressed RBDs (1 µg/ml) reacted with the anti-RBD mAbs (2.2 µg/ml), RBD219-WT and RBD219-N1 exhibited the strongest binding to all tested mAbs. The reactivity of RBD193-N3 with Conf IV mAb 35B5, Conf VI mAb 19B2, and linear mAb 17H9 was considerably lower than that of the other wild-type and mutant RBDs. Decrease in mAb concentration to 0.25 µg/ml did not significantly affect the binding of RBD219-N1 or RBD219-WT to all five conformational mAbs, whereas their reactivity to the linear anti-RBD mAb 17H9 was significantly decreased (). These data suggest that the deglycosylated RBD219-N1 protein, like RBD219-WT, was able to maintain conformation and antigenicity, despite the deletion of the N1 glycosylation site.
Figure 6. Detection of the reactivity of yeast-expressed RBD proteins with RBD-specific mAbs by ELISA. The mAbs specific for the conformational epitopes (24H8 (Conf I), 19B2 (VI), 35B5 (Conf IV), 33G4 (Conf V), 31H12 (Conf II)), and linear epitopes (17H9) of SARS-CoV RBD were tested at 2.2 µg/ml (A) or 0.25 µg/ml (B), respectively. Wild-type SARS-CoV RBD protein expressed in 293T cells (RBD193-WT)Citation11 was included as the positive control. The data in (A) and (B) are expressed as the mean ± standard deviation (SD) of duplicate wells.
Functionality of yeast-expressed RBD proteins
The functionality of yeast-expressed RBD proteins was further confirmed based on the ability of these proteins to bind to ACE2, the receptor of SARS-CoV, in either cell-associated or soluble form (sACE2). To establish the binding of these proteins to the cell-associated ACE2, a test protein (20 µg/each) was first mixed with ACE2/293T cells in the presence of linear anti-RBD mAb 17H9Citation14 and protein A and G beads, followed by western blot using a specific antibody against ACE2. As shown in , one clear band corresponding to the size of ACE2 was observed in the cell lysates pulled down with the respective RBD proteins; all reacted strongly with the ACE2-specific mAb, although the RBD193-N3 mixture showed weaker reaction. These results suggest that the yeast-expressed RBD proteins are able to bind efficiently to the cell-surface receptor ACE2. As expected, no band (binding) was shown in the line of control protein (MERS-CoV RBD) ().
Figure 7. Binding of SARS-CoV RBD proteins with cell-associated ACE2 (ACE2/293T cells) or sACE2. Binding of RBD proteins (20 µg/each) with ACE2/293T cells (A) or sACE2 (20 µg) (B and C) was detected by western blot using goat anti-ACE2 mAb (0.2 µg/ml) or anti-RBD of SARS-CoV mAb (33G4, 1 µg/ml). A recombinant protein containing RBD of Middle East respiratory syndrome coronavirus (MERS-CoV)Citation28 was included as the negative control (control).
We further detected the binding of the RBD proteins with sACE2 by mixing equal concentration of an RBD protein with sACE2 (containing 6 × His tag) in the presence of Ni-NTA beads, followed by western blot as above. As shown in , two clear bands corresponding to the size of sACE2 and respective RBD proteins were revealed in the pulled-down samples, while only one band corresponding to the size of sACE2 or respective RBD proteins, was shown in the samples containing either sACE2 or RBD protein only, all of which reacted strongly with antibodies against ACE2 or SARS-CoV RBD (33G4), respectively. However, only one band corresponding to the size of sACE2 was shown in the control containing RBD protein of MERS-CoV plus sACE2, or sACE2 only (). These results confirm the specific binding of these yeast-expressed RBDs with SARS-CoV’s receptor ACE2, indicating that all RBD proteins with or without mutations maintain sufficient functionality.
Yeast-expressed RBD proteins of SARS-CoV elicited a robust systemic humoral immune response
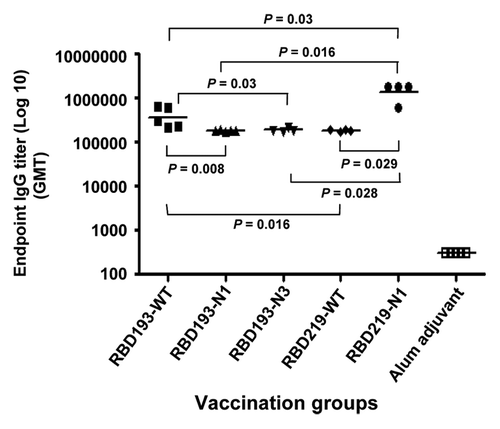
To compare the immunogenicity of yeast-expressed RBD proteins, we immunized mice using these proteins and analyzed the IgG antibody responses in mouse sera collected at 10 d post-last vaccination. As shown in , all yeast-expressed RBD proteins were able to induce sufficient IgG antibody responses against RBD219-WT. Particularly, RBD219-N1 induced significantly higher IgG antibody responses against RBD219-WT than other RBD proteins, including RBD193-WT, RBD193-N1, RBD193-N3, and RBD219-WT, with the geometric mean titers of 1.4 × 106 (RBD219-N1), 3.5 × 105 (RBD193-WT), 1.8 × 105 (RBD193-N1), 1.9 × 105 (RBD193-N3), and 1.8 × 105 (RBD219-WT), respectively. By comparison, SARS-CoV RBD193-WT induced significantly higher IgG antibody responses against RBD219-WT than RBD193-N1, RBD193-N3, and RBD219-WT, respectively. No significant differences were observed between SARS-CoV RBD193-N1, RBD193-N3, or RBD219-WT. The control mice, which were immunized with alum plus PBS, only showed background antibody responses (). These results suggest that RBD219-N1 exhibited the highest immunogenicity, eliciting, in turn, the strongest RBD-specific antibody responses in the vaccinated mice.
Figure 8. Detection of SARS-CoV RBD-specific IgG antibody by ELISA in the vaccinated mouse sera. Sera collected at 10 d post-last vaccination were used for the test. Alum adjuvant plus PBS was used as the control. The data are presented as geometric mean titers (GMT) of five mice per group. P values indicate significant differences between different vaccination groups.
Yeast-expressed SARS-CoV RBD proteins induced in vaccinated mice comparable titers of neutralizing antibody against the SARS-CoV
To compare the ability of the yeast-expressed SARS-CoV RBD proteins to elicit neutralizing antibodies, the sera collected from vaccinated mice 10 d after the last vaccination was tested using a SARS pseudovirus-based neutralization assay. As shown in , immunization with RBD193-WT, RBD219-WT, or RBD219-N1 uniformly elicited potent neutralizing antibody responses, with the neutralizing antibody titers around 4 × 104, respectively, which were significantly stronger than those induced by either RBD193-N1 or RBD193-N3, with the neutralizing antibody titers of 4.3 × 103 and 1.4 × 104, respectively. As expected, the alum adjuvant plus PBS control induced no neutralizing antibody response against the SARS pseudovirus ().
Figure 9. Detection of neutralizing antibodies in sera of vaccinated mice against pseudotyped and live SARS-CoV infection. Sera collected at 10 d post-last vaccination were used for the test. Alum adjuvant plus PBS was used as the control. (A) Titers of neutralizing antibodies against SARS pseudovirus. The data are expressed as NT50 and are presented as mean ± SD of five mice per group. (B) Titers of neutralizing antibodies against live SARS-CoV infection. NT50 was presented as mean ± SD of five mice per group. P values indicate significant differences between different vaccination groups.
To further confirm the functionality of the induced neutralizing antibodies, a live SARS-CoV-based neutralization assay was then performed. As shown in , immunization with RBD219-N1 resulted in significantly higher titers of neutralizing antibody responses against live SARS-CoV infection than those elicited by RBD193-WT, RBD193-N1, RBD193-N3, or even RBD219-WT, with the neutralizing antibody titers reaching 4.5 × 103 (RBD219-N1), 2.3 × 103 (RBD193-WT), 2.5 × 102 (RBD193-N1), 1.6 × 103 (RBD193-N3), and 2.2 × 103 (RBD219-WT), respectively. While no significant difference was seen between RBD193-WT and RBD219-WT groups, the titers of neutralizing antibodies against live SARS-CoV infection induced by RBD193-N1 and RBD193-N3 were significantly lower than those induced by RBD193-WT. Similarly, the alum control group did not induce neutralizing antibodies against live SARS-CoV (). The above data further confirm that RBD219-N1 possessed the strongest immunogenicity among RBD proteins tested in inducing potent neutralizing antibody responses against live SARS-CoV in vaccinated animals.
Materials and Methods
Cloning and expression of RBDs in yeast Pichia pastoris
The DNAs encoding the 193-aa (RBD193, residues 318–510) and 219-aa (RBD219, residues 318–536) of SARS-CoV RBD were codon-optimized based on yeast codon usage preference and synthesized by GenScript, followed by subcloning into Pichia secretory expression vector pPICZαA (Invitrogen) using EcoRI/XbaI restriction sites. The correct insert sequences and reading frames of recombinant plasmids were confirmed by double-stranded sequencing using vector flanking primers α-factor and 3′AOX-1. The recombinant plasmid DNAs were then transformed into Pichia pastoris X-33 by electroporation. The expressions of recombinant RBD193 and RBD219 were induced with 0.5% methanol at 30 °C for 72 h, and the highest expression clone was chosen to make seed stock in 20% glycerol as described previously.Citation16
Because high glycosylation of the RBD193/RBD219-wild-type (WT) expressed in yeast could cause yield and reproducibility problems, the three asparagines in the RBD sequence responsible for N-glycosylation were removed or mutated to make deglycosylated forms as follows: N1: deletion of the 1st Asn (N-1), the 1st glycosylation site; N2: mutation of the 2nd N-glycosylated Asn (N-13) to Ser in addition to the N1 deletion; and N3: mutation of the 3rd N-glycosylated N-30 to Ala, in addition of N1 and N2 deletion/mutation (). The expression and glycosylation level of the recombinant RBD193-WT and RBD219-WT, and their deglycosylated forms was confirmed by SDS-PAGE and western blot with anti-RBD mAb 33G4.Citation14
Glycosidase Assay
In order to determine whether the expressed recombinant RBD193-WT is glycosylated, yeast-expressed RBD193-WT was digested with Peptide-N-Glycosidase F (PNGase F) (New England Biolabs (NEB)). Briefly, 10 µl of the RBD193-WT/pPICZαA/P. pastoris culture induced with 0.5% methanol for 72 h were mixed with 1 µl of denaturing buffer (NEB) in a 1.5 ml tube and denatured at 100 °C for 10 min. Then 2 µl of G7 buffer, 2 µl of 10% NP40 (all came with PNGase F), 1 µl of N-PNGase F, and 5 µl of deionized water were added in the tube. The mixture was then incubated at 37 °C for an hour. The removal of glycans was confirmed by SDS-PAGE, followed by western blot using anti-RBD mAb 33G4.
Optimization of induction conditions by pH and detergent
Seed of RBD193-WT/pPICZαA/P. pastoris X-33 was grown in 5 ml of Buffered Glycerol-complex Medium (BMGY) at 225 rpm at 30 °C overnight until the OD600 reached 2–6. The expression of recombinant RBD193-WT was induced in 10 ml Buffered Methanol-complex Medium (BMMY) (starting OD600 = 1.0) containing 0.5% methanol with different pH of 5.2, 6.0, 7.5, and 8.0. EMPIGEN ® BB Detergent was added into the culture (pH 6.0) at final concentration of 0.01% and 0.05% in order to determine if the detergent was able to disrupt any possible aggregation of the expressed recombinant protein. The induction was continued for 72 h. The expression, yield, and integrity of the expressed recombinant RBD193 in different culture media were identified by western blot using anti-RBD mAb 33G4.
Fermentation and purification
To scale-up the expression of recombinant RBDs in yeast, the constructs of RBD193-N1, RBD193-N3, RBD219-WT, and RBD219-N1 in pPICZαA/P. pastoris X33 were fermented in 5 L fermentation as described previously.Citation1 Briefly, the seed stocks of each construct were used to inoculate 1 L of Buffered Minimal Glycerol (BMG) medium and cultured overnight at 30 °C with shaking at 225 rpm until the OD600 reached ~10.0. 110 ml of this culture were used to inoculate 2.5 L of sterile BSM in the fermenter containing 3.5 ml/L of PTM1 trace elements and 3.5 ml/L of 0.02% d-Biotin. Fermentation was initiated at 30 °C, and the initial pH was set at 5.0. Gas and agitation were adjusted to maintain dissolved oxygen (DO) at 30%. Upon exhaustion of glycerol during the batch phase (DO spike), methanol was pumped in from 0.8 ml/L/h to 10 ml/L/h over a 6–8 h period, the pH was adjusted to 6.0 using 14% ammonium hydroxide, and the induction was maintained at 26 °C for 75 h. After fermentation, the culture was harvested by centrifugation at 7000 rpm for 30 min at 4 °C and filtered through a 0.22 PM bottle top filtration unit. The expression yield of recombinant RBDs in the fermented culture was determined by SDS-PAGE and densitometry. To purify RBD193N1, RBD219-WT, and RBD219-N1, one part of fermentation supernatant was diluted with 2 parts of 30 mM Tris and 3 M ammonium sulfate, at pH 8.0, and then loaded to HiTrap Butyl Sepharose HP at the flow rate of 1.5 ml/min, followed by washing with 30 mM Tris and 2 M ammonium sulfate to remove the unbound proteins. The bound RBD protein was eluted with gradient ammonium sulfate starting from 2 M. The fractions containing target protein were pooled together, concentrated, and further purified with Toyopearl HW55S size exclusion column in order to eliminate the remaining contaminants. To purify RBD193-N3, the fermented culture supernatant was chromatographed using an anion exchange Q Sepharose XL column. The flow-through was collected, concentrated and, like other RBD proteins, further purified with Toyopearl HW55S size exclusion column. The purity of the yeast-expressed recombinant RBDs and the positive control (RBD193-WT/293T) was confirmed by SDS-PAGE and Western Blot using anti-RBD mAbs 33G4, 35B5, 24H8, and 31H12 developed in our laboratory.Citation14
Animals
Four- to six-week-old female BALB/c mice were used for the study and housed in the animal facility of the New York Blood Center. The animal studies were performed in accordance with the recommendations for the Care and Use of Laboratory Animals of the National Institutes of Health. The animal protocol was approved by the Committee on the Ethics of Animal Experiments of the New York Blood Center (Permit Number: 194.14).
Mouse vaccination and sera collection
The immunization protocols were performed as previously described with some modifications.Citation11,Citation12,Citation15 Briefly, mice were subcutaneously (s.c.) immunized with yeast-expressed recombinant RBD proteins (RBD193-N1, RBD193-N3, RBD219-N1, or RBD219-WT) (20 μg/mouse) formulated with Alhydrogel 2% (aluminum hydroxide gel, hereinafter alum) adjuvant (InvivoGen). A mammalian cell 293T-expressing SARS-CoV RBD protein (RBD193-WT) [11] and PBS were used as positive and negative controls, respectively. Immunized mice were boosted twice with the same alum-formulated immunogen (10 μg/mouse) at 21-d intervals. Mouse sera were collected before immunization and 10 d after each vaccination to measure humoral IgG antibody responses and neutralizing antibodies.
ELISA
ELISA was used to verify the conformation of the yeast-expressed SARS-CoV RBD proteins to RBD-specific mAbs developed in our laboratory.Citation14 Briefly, 96-well ELISA plates were respectively pre-coated with each yeast-expressed RBD protein (1 µg/ml) overnight at 4 °C and blocked with 2% non-fat milk for 2 h at 37 °C. A series of conformation-dependent mAbs, including 24H8 (Conf I), 31H12 (Conf II), 35B5 (Conf IV), 33G4 (Conf V), 19B2 (Conf VI), and linear-dependent mAb 17H9,Citation14 were added to the plates and incubated for 1 h at 37 °C, followed by four washes. Bound antibodies were reacted with horseradish peroxidase (HRP)-conjugated anti-mouse IgG (1:3000, Invitrogen) for 1 h at 37 °C. After four washes, the substrate 3,3′,5,5′-tetramethylbenzidine (TMB) (Zymed) was added to the plates, and the reaction was stopped by adding 1 N H2SO4. The absorbance at 450 nm (A450) was measured with an ELISA plate reader (Tecan).
In addition, the reactivity of polyclonal antibodies in the collected mouse sera against the expressed RBD was also measured by ELISA using our previously described protocols with some modifications.Citation11,Citation12,Citation15 Briefly, 96-well ELISA plates were respectively pre-coated with yeast-expressed RBD219-WT protein (1 µg/ml) overnight at 4 °C. After blocking and then incubating with serially diluted mouse sera, bound IgG antibody was detected by using HRP-conjugated anti-mouse IgG (1:2000), followed by the same protocol as described above.
Pull-down binding assay
The binding of yeast-expressed recombinant SARS-CoV RBD proteins to cell-associated receptor ACE2 or soluble ACE2 (sACE2) (R&D Systems) was performed by pull-down assay using previously described protocols with some modifications.Citation17 Briefly, the lysates of 293T cells expressing ACE2 (ACE2/293T) or sACE2 were respectively incubated with recombinant SARS-CoV RBD proteins plus 17H9 linear mAb.Citation14 and Protein A and G (for cell-associated ACE2) or Ni-NTA affinity column (for sACE2). After rotation at 4 °C overnight, the supernatant of the mixture was removed by centrifugation. After washing 3 times with PBS, the pellet with binding proteins was boiled for 10 min and the supernatants were subjected to SDS-PAGE and western blot as described below.
SDS-PAGE and western blot
SDS-PAGE and western blot were performed using previously described protocols with some modifications.Citation18,Citation19 Briefly, the pull-down proteins or the expressed recombinant RBDs were subjected to SDS-PAGE and transferred to a nitrocellulose membrane, which was blocked with 5% non-fat milk in PBS with 0.05% Tween-20 (PBST) at 4 °C overnight, followed by successive incubation with goat anti-ACE2 mAb (1 μg/ml) or anti-RBD mAb (0.2 μg/ml), respectively, and HRP-labeled anti-goat IgG (1:1,000, R&D Systems) for 1 h at room temperature. Signals were visualized with ECL western blot substrate reagents (GE Healthcare) and Amersham Hyperfilm (GE Healthcare).
Pseudovirus neutralization assay
Neutralizing antibody titers of recombinant RBD-immunized mouse sera were measured using a pseudovirus neutralization assay as previously described with some modifications.Citation11,Citation15 Briefly, 293T cells were co-transfected with a plasmid encoding SARS-CoV S protein and a plasmid encoding Env-defective, luciferase-expressing HIV-1 genome (pNL4–3.luc.RE), using the calcium phosphate method. The culture supernatant was harvested 72 h post-transfection and used for single-cycle infection of 293T cells expressing SARS-CoV’s receptor ACE2 (ACE2/293T). The cells were seeded in 96-well culture plates at 104/well and incubated at 37 °C for 4–6 h to form a monolayer. Serial 2-fold diluted mouse sera were mixed with SARS-CoV pseudovirus at 37 °C for 1 h and then transferred to the monolayer cells. After incubation for 72 h, relative luciferase activity was measured by Ultra 384 luminometer (Tecan). SARS pseudovirus neutralization was calculated and expressed as NT50.
Live virus-based neutralization assay
The neutralizing titers of recombinant RBD-immunized mouse sera were further measured using a live virus-based neutralization assay as previously described with some modifications.Citation20,Citation21 Briefly, serial 2-fold diluted mouse sera were mixed with ~100 infectious SARS-CoV at 37 °C for 1 h and then added to the monolayer of Vero E6 cells in duplicate. Cytopathic effect (CPE) in each well was observed daily and recorded on day 3 post-infection. The neutralizing titers were reported as the reciprocal of the highest dilution of serum that completely prevented CPE in at least 50% of the wells (NT50).
Discussion
The RBD of SARS-CoV S protein contains multiple conformation-dependent epitopes that induce potent neutralizing antibody responses against a broad spectrum of SARS-CoV strains, thus serving as an important target for developing SARS vaccines.Citation14,Citation22-Citation24 We have previously demonstrated that the recombinant protein containing 193 amino acids of the RBD domain fused with an Fc tag (RBD193-Fc) induced highly potent neutralizing antibody responses and protective immunity in vaccinated animals.Citation11,Citation12,Citation15 To eliminate potential adverse effects in humans, possibly induced by the Fc fragment in these vaccine candidates, we have successively expressed several RBD proteins without the Fc tag in different expression systems, including mammalian 293T and CHO-K1 cells, Sf9 inset cells and E. coli. It was shown that most of these RBD proteins without the Fc tag could still induce strong neutralizing antibody response and protection against SARS-CoV challenge in the vaccinated animals.Citation11,Citation12,Citation15 These findings suggested that the RBD protein on itself can elicit potent neutralizing antibody response and protect animals and we assume also people from SARS-CoV infection.
SARS-CoV S-RBD (residues 318–510) contains three N-linked glycosylation sites at the N-1, N-13 and N-40 positions.Citation10 While a mammalian cell-expressed RBD protein with high glycosylation induced significant neutralizing activity, an E. coli-expressed RBD protein without the glycans was able to react with the RBD-specific, conformation-dependent mAbs, and also induced substantial neutralizing antibody responses in the vaccinated animals,Citation11 suggesting that recombinant RBD proteins without the glycans can still maintain their essential antigenicity and immunogenicity when used as a candidate vaccine.
The methylotrophic yeast Pichia pastoris has been widely used for heterogeneous protein expression for pharmaceutical and vaccine applications owing to its ability to: 1) produce large amounts of protein in defined media absent animal-derived growth factors; and 2) offer easy scale-up at low cost.Citation16,Citation25 As an eukaryotic expression system, yeast is capable of many post-translational modifications, such as proteolytic processing, folding, disulfide bond formation and glycosylation, which may be necessary for the functions of the expressed proteins. However, different from mammalian cells, Pichia pastoris frequently adds monosaccharide to expressed protein to become hyperglycosylated.Citation26 The extent and linkage of residues present in hyperglycosylated glycans vary, depending on yeast strain and cell culture conditions,Citation27 causing, as a consequence, concerns regarding expression yield, reproducibility of homogeneous products and quality control.Citation28 Because of these concerns, the three asparagines responsible for N-glycosylation in the RBD sequence were removed or mutated to make deglycosylated RBD forms.
In the present study, we expressed two wild-type RBDs with different lengths (RBD193-WT and RBD219-WT) and their deglycosylated mutants (RBD193-N1, RBD193-N2, RBD193-N3; RBD219-N1, RBD219-N2, RBD219-N3) by mutating or deleting residues at the corresponding glycosylation sites (). Their antigenicity, functionality, and immunogenicity were then compared. SDS-PAGE analysis revealed that both yeast-expressed RBD193-WT and RBD219-WT migrated as a smear with apparent molecular weights higher than their calculated M.W., suggesting that these recombinant RBD-WTs were highly glycosylated or aggregated (). After digestion of RBD193-WT with N-glycosidase PNGase F, the high M.W. smear disappeared, and the size of RBD193-WT returned to the expected M.W. (), confirming that the high M.W. smear was from hyperglycosylation of the yeast-expressed RBD, rather than any aggregation. When some of the N-linked glycosylation sites in RBD193-WT and RBD219-WT were deleted or mutated (N1, N2, and N3), the apparent M.W. of recombinant RBDs was reduced accordingly (). Therefore, these deglycosylated forms have allowed us to accurately and reproducibly control the expression process during scale-up production and quality control testing. Notably, the expression yield of the recombinant RBDs with deleted or mutated N-linked glycosylation sites was reduced compared with that of their corresponding wild-type RBDs (); therefore, pointing to the need to find a workable balance between expression yield and glycosylation level during product development.
Importantly, RBD219-N1 exhibited a lower glycosylation level without obvious compromised expression yield when compared with the wild-type form (). After two-step purification, including hydrophobic interaction (Butyl HP) chromatography and size exclusion column, most of the glycosylation species and the host protein contaminations were removed ( and ). Consequently, we selected the recombinant RBD219-N1 protein for further evaluation. We found that RBD219-N1 was highly recognized by the conformational anti-RBD mAbsCitation14,Citation24 using two separate methods; western blot (), and ELISA (), further validating the ability of this deglycosylated protein to maintain antigenicity that is equal to that of the wild-type protein. Moreover, like the wild-type RBD, RBD219-N1 bound very well to the cell-associated and soluble receptor ACE2 (), further confirming that this yeast-expressed RBD variant maintains its functionality.
We then compared the immunogenicity of RBD219-N1 with wild-type RBDs and some of the other deglycosylated proteins. We found that RBD219-N1 induced remarkably higher SARS-CoV RBD-specific IgG antibody responses than the wild-type RBDs, RBD219-WT, and RBD193-WT, and the other deglycosylated proteins, such as RBD193-N1 and RBD193-N3. However, RBD193-N1 and RBD193-N3 exhibited relatively lower antigenicity and immunogenicity than RBD193-WT (, and ). Among all the tested yeast-expressed RBDs, RBD219-N1 elicited the highest titers of neutralizing antibodies against the pseudotyped SARS-CoV and live SARS-CoV infection (). Why do RBD219-N1 and RBD193-N1 induce high and low neutralization titers, respectively? Crystal structure of SARS-CoV RBD complexed with ACE2 shows that all the glycosylation sites in the RBD are away from the ACE2-binding sites, and thus deglycosylation of the RBD should not interfere with ACE2 binding.Citation29 However, based on the most complete structural model of the SARS-CoV RBD, the C-terminal region of the RBD after residue 512 is disordered but close to the N-terminus of the RBD,Citation30 and thus the glycosylation site at the N-terminus may have steric interference with the C-terminal region of the RBD. Therefore, removing the glycosylation site at the N-terminus may help stabilize the overall structure of RBD219 and lead to high immunization titers. In contrast, deglycosylation of RBD193-N1 and/or two other glycosylation sites in the RBD of SARS-CoV may cause the immune system to lose focus on the ACE2-binding region and lead to reduced neutralizing titers.Citation31,Citation32
Further evaluation of the RBD219-N1 vaccine candidate in vivo for efficacy, protecting animals from SARS-CoV challenge, is under investigation. According to our previous experience, an RBD-based vaccine candidate that can induce NT50 (50% neutralizing antibody titer) >1,000 of neutralizing antibody titers against the live SARS-CoV is expected to fully protect animals from infection by the virus.Citation11,Citation15 Our present data showed that the titer of neutralizing antibodies induced by RBD219-N1 in vaccinated mice against live SARS-CoV was >2,000 (), demonstrating its prospect to efficiently protect immunized animals from an SARS-CoV challenge.
In conclusion, the yeast-expressed RBD without any extra tag but with the glycosylation site at N1 deleted, RBD219-N1, can exhibit lower glycosylation levels and higher expression yield, and induced stronger RBD-specific antibody responses and more potent neutralizing antibodies against both pseudotyped and live SARS-CoV, pointing to its potential use for further development as an effective and safe subunit vaccine against SARS-CoV in humans.
| Abbreviations: | ||
| ACE2 | = | angiotensin-converting enzyme 2 |
| DO | = | dissolved oxygen |
| mAb | = | monoclonal antibody |
| RBD | = | receptor-binding domain |
| SARS | = | severe acute respiratory syndrome |
| SARS-CoV | = | SARS coronavirus |
| S | = | spike |
| WT | = | wild-type |
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Funding
This study was supported by grant from the National Institutes of Health (R01AI098775).
Acknowledgments
We thank Ms Diane Niño (the project manager) for coordinating the study. We also thank Dr Fang Li at Department of Pharmacology, University of Minnesota Medical School for the helpful discussion and comments on the structural analysis of the SARS-CoV RBD.
References
- Du L, He Y, Zhou Y, Liu S, Zheng BJ, Jiang S. The spike protein of SARS-CoV--a target for vaccine and therapeutic development. Nat Rev Microbiol 2009; 7:226 - 36; http://dx.doi.org/10.1038/nrmicro2090; PMID: 19198616
- Peiris JSM, Lai ST, Poon LLM, Guan Y, Yam LY, Lim W, Nicholls J, Yee WK, Yan WW, Cheung MT, et al, SARS study group. Coronavirus as a possible cause of severe acute respiratory syndrome. Lancet 2003; 361:1319 - 25; http://dx.doi.org/10.1016/S0140-6736(03)13077-2; PMID: 12711465
- Zhong NS, Zheng BJ, Li YM, Poon, Xie ZH, Chan KH, Li PH, Tan SY, Chang Q, Xie JP, et al. Epidemiology and cause of severe acute respiratory syndrome (SARS) in Guangdong, People’s Republic of China, in February, 2003. Lancet 2003; 362:1353 - 8; http://dx.doi.org/10.1016/S0140-6736(03)14630-2; PMID: 14585636
- Jiang S, Bottazzi ME, Du L, Lustigman S, Tseng CT, Curti E, Jones K, Zhan B, Hotez PJ. Roadmap to developing a recombinant coronavirus S protein receptor-binding domain vaccine for severe acute respiratory syndrome. Expert Rev Vaccines 2012; 11:1405 - 13; http://dx.doi.org/10.1586/erv.12.126; PMID: 23252385
- Perlman S, Dandekar AA. Immunopathogenesis of coronavirus infections: implications for SARS. Nat Rev Immunol 2005; 5:917 - 27; http://dx.doi.org/10.1038/nri1732; PMID: 16322745
- Bolles M, Deming D, Long K, Agnihothram S, Whitmore A, Ferris M, Funkhouser W, Gralinski L, Totura A, Heise M, et al. A double-inactivated severe acute respiratory syndrome coronavirus vaccine provides incomplete protection in mice and induces increased eosinophilic proinflammatory pulmonary response upon challenge. J Virol 2011; 85:12201 - 15; http://dx.doi.org/10.1128/JVI.06048-11; PMID: 21937658
- Castilow EM, Olson MR, Varga SM. Understanding respiratory syncytial virus (RSV) vaccine-enhanced disease. Immunol Res 2007; 39:225 - 39; http://dx.doi.org/10.1007/s12026-007-0071-6; PMID: 17917067
- Tseng CT, Sbrana E, Iwata-Yoshikawa N, Newman PC, Garron T, Atmar RL, Peters CJ, Couch RB. Immunization with SARS coronavirus vaccines leads to pulmonary immunopathology on challenge with the SARS virus. PLoS One 2012; 7:e35421; http://dx.doi.org/10.1371/journal.pone.0035421; PMID: 22536382
- Jaume M, Yip MS, Kam YW, Cheung CY, Kien F, Roberts A, Li PH, Dutry I, Escriou N, Daeron M, et al. SARS CoV subunit vaccine: antibody-mediated neutralisation and enhancement. Hong Kong Med J 2012; 18:Suppl 2 31 - 6; PMID: 22311359
- Wong SK, Li W, Moore MJ, Choe H, Farzan M. A 193-amino acid fragment of the SARS coronavirus S protein efficiently binds angiotensin-converting enzyme 2. J Biol Chem 2004; 279:3197 - 201; http://dx.doi.org/10.1074/jbc.C300520200; PMID: 14670965
- Du L, Zhao G, Chan CC, Sun S, Chen M, Liu Z, Guo H, He Y, Zhou Y, Zheng BJ, et al. Recombinant receptor-binding domain of SARS-CoV spike protein expressed in mammalian, insect and E. coli cells elicits potent neutralizing antibody and protective immunity. Virology 2009; 393:144 - 50; http://dx.doi.org/10.1016/j.virol.2009.07.018; PMID: 19683779
- Du L, Zhao G, Chan CC, Li L, He Y, Zhou Y, Zheng BJ, Jiang S. A 219-mer CHO-expressing receptor-binding domain of SARS-CoV S protein induces potent immune responses and protective immunity. Viral Immunol 2010; 23:211 - 9; http://dx.doi.org/10.1089/vim.2009.0090; PMID: 20374001
- Chen Z, Zhang L, Qin C, Ba L, Yi CE, Zhang F, Wei Q, He T, Yu W, Yu J, et al. Recombinant modified vaccinia virus Ankara expressing the spike glycoprotein of severe acute respiratory syndrome coronavirus induces protective neutralizing antibodies primarily targeting the receptor binding region. J Virol 2005; 79:2678 - 88; http://dx.doi.org/10.1128/JVI.79.5.2678-2688.2005; PMID: 15708987
- He Y, Lu H, Siddiqui P, Zhou Y, Jiang S. Receptor-binding domain of severe acute respiratory syndrome coronavirus spike protein contains multiple conformation-dependent epitopes that induce highly potent neutralizing antibodies. J Immunol 2005; 174:4908 - 15; PMID: 15814718
- Du L, Zhao G, Li L, He Y, Zhou Y, Zheng BJ, Jiang S. Antigenicity and immunogenicity of SARS-CoV S protein receptor-binding domain stably expressed in CHO cells. Biochem Biophys Res Commun 2009; 384:486 - 90; http://dx.doi.org/10.1016/j.bbrc.2009.05.003; PMID: 19422787
- Goud GN, Zhan B, Ghosh K, Loukas A, Hawdon J, Dobardzic A, Deumic V, Liu S, Dobardzic R, Zook BC, et al. Cloning, yeast expression, isolation, and vaccine testing of recombinant Ancylostoma-secreted protein (ASP)-1 and ASP-2 from Ancylostoma ceylanicum. J Infect Dis 2004; 189:919 - 29; http://dx.doi.org/10.1086/381901; PMID: 14976610
- Du L, Zhao G, Kou Z, Ma C, Sun S, Poon VK, Lu L, Wang L, Debnath AK, Zheng BJ, et al. Identification of a receptor-binding domain in the S protein of the novel human coronavirus Middle East respiratory syndrome coronavirus as an essential target for vaccine development. J Virol 2013; 87:9939 - 42; http://dx.doi.org/10.1128/JVI.01048-13; PMID: 23824801
- Du L, Zhao G, Sun S, Zhang X, Zhou X, Guo Y, Li Y, Zhou Y, Jiang S. A critical HA1 neutralizing domain of H5N1 influenza in an optimal conformation induces strong cross-protection. PLoS One 2013; 8:e53568; http://dx.doi.org/10.1371/journal.pone.0053568; PMID: 23320093
- Du L, Leung VH, Zhang X, Zhou J, Chen M, He W, Zhang HY, Chan CC, Poon VK, Zhao G, et al. A recombinant vaccine of H5N1 HA1 fused with foldon and human IgG Fc induced complete cross-clade protection against divergent H5N1 viruses. PLoS One 2011; 6:e16555; http://dx.doi.org/10.1371/journal.pone.0016555; PMID: 21304591
- Du L, Zhao G, Lin Y, Sui H, Chan C, Ma S, He Y, Jiang S, Wu C, Yuen KY, et al. Intranasal vaccination of recombinant adeno-associated virus encoding receptor-binding domain of severe acute respiratory syndrome coronavirus (SARS-CoV) spike protein induces strong mucosal immune responses and provides long-term protection against SARS-CoV infection. J Immunol 2008; 180:948 - 56; PMID: 18178835
- Du L, Zhao G, Lin Y, Chan C, He Y, Jiang S, Wu C, Jin DY, Yuen KY, Zhou Y, et al. Priming with rAAV encoding RBD of SARS-CoV S protein and boosting with RBD-specific peptides for T cell epitopes elevated humoral and cellular immune responses against SARS-CoV infection. Vaccine 2008; 26:1644 - 51; http://dx.doi.org/10.1016/j.vaccine.2008.01.025; PMID: 18289745
- He Y, Zhou Y, Liu S, Kou Z, Li W, Farzan M, Jiang S. Receptor-binding domain of SARS-CoV spike protein induces highly potent neutralizing antibodies: implication for developing subunit vaccine. Biochem Biophys Res Commun 2004; 324:773 - 81; http://dx.doi.org/10.1016/j.bbrc.2004.09.106; PMID: 15474494
- He Y, Zhu Q, Liu S, Zhou Y, Yang B, Li J, Jiang S. Identification of a critical neutralization determinant of severe acute respiratory syndrome (SARS)-associated coronavirus: importance for designing SARS vaccines. Virology 2005; 334:74 - 82; http://dx.doi.org/10.1016/j.virol.2005.01.034; PMID: 15749124
- He Y, Li J, Li W, Lustigman S, Farzan M, Jiang S. Cross-neutralization of human and palm civet severe acute respiratory syndrome coronaviruses by antibodies targeting the receptor-binding domain of spike protein. J Immunol 2006; 176:6085 - 92
- Punt PJ, van Biezen N, Conesa A, Albers A, Mangnus J, van den Hondel C. Filamentous fungi as cell factories for heterologous protein production. Trends Biotechnol 2002; 20:200 - 6; http://dx.doi.org/10.1016/S0167-7799(02)01933-9; PMID: 11943375
- Dean N. Asparagine-linked glycosylation in the yeast Golgi. Biochim Biophys Acta 1999; 1426:309 - 22; http://dx.doi.org/10.1016/S0304-4165(98)00132-9; PMID: 9878803
- Shibata N, Ichikawa T, Tojo M, Takahashi M, Ito N, Okubo Y, Suzuki S. Immunochemical study on the mannans of Candida albicans NIH A-207, NIH B-792, and J-1012 strains prepared by fractional precipitation with cetyltrimethylammonium bromide. Arch Biochem Biophys 1985; 243:338 - 48; http://dx.doi.org/10.1016/0003-9861(85)90511-9; PMID: 3002275
- Hopkins D, Gomathinayagam S, Rittenhour AM, Du M, Hoyt E, Karaveg K, Mitchell T, Nett JH, Sharkey NJ, Stadheim TA, et al. Elimination of β-mannose glycan structures in Pichia pastoris. Glycobiology 2011; 21:1616 - 26; http://dx.doi.org/10.1093/glycob/cwr108; PMID: 21840970
- Li F, Li W, Farzan M, Harrison SC. Structure of SARS coronavirus spike receptor-binding domain complexed with receptor. Science 2005; 309:1864 - 8; http://dx.doi.org/10.1126/science.1116480; PMID: 16166518
- Prabakaran P, Gan J, Feng Y, Zhu Z, Choudhry V, Xiao X, Ji X, Dimitrov DS. Structure of severe acute respiratory syndrome coronavirus receptor-binding domain complexed with neutralizing antibody. J Biol Chem 2006; 281:15829 - 36; http://dx.doi.org/10.1074/jbc.M600697200; PMID: 16597622
- Zakhartchouk AN, Sharon C, Satkunarajah M, Auperin T, Viswanathan S, Mutwiri G, Petric M, See RH, Brunham RC, Finlay BB, et al. Immunogenicity of a receptor-binding domain of SARS coronavirus spike protein in mice: implications for a subunit vaccine. Vaccine 2007; 25:136 - 43; http://dx.doi.org/10.1016/j.vaccine.2006.06.084; PMID: 16919855
- Trujillo JD, Kumpula-McWhirter NM, Hötzel KJ, Gonzalez M, Cheevers WP. Glycosylation of immunodominant linear epitopes in the carboxy-terminal region of the caprine arthritis-encephalitis virus surface envelope enhances vaccine-induced type-specific and cross-reactive neutralizing antibody responses. J Virol 2004; 78:9190 - 202; http://dx.doi.org/10.1128/JVI.78.17.9190-9202.2004; PMID: 15308714
