Abstract
Differential use of cellular and molecular components shapes immune responses, but understanding of how these are regulated to promote defense and health during infections is still incomplete. Examples include signaling from members of the Janus activated kinase-signal transducer and activator of transcription (JAK-STAT) cytokine family. Following receptor stimulation, individual JAK-STAT cytokines have preferences for particular key STAT molecules to lead to specific cellular responses. Certain of these cytokines, however, can conditionally activate alternative STATs as well as elicit pleiotropic and paradoxical effects. Studies examining basal and infection conditions are revealing intrinsic and induced cellular differences in various intracellular STAT concentrations to control the biological consequences of cytokine exposure. The system can be likened to changing partners at a dance based on competition and relative availability, and sets a framework for understanding the particular conditions promoting subset biological functions of cytokines as needed during evolving immune responses to infections.
Introduction
The immune system has a complex but flexible network of cellular and molecular components to deal with the constraints of limited genetic space in responding to almost limitless types of infectious organisms, i.e., viral, bacterial, fungal and parasitic variants. These components are broadly divided into innate and antigen-specific adaptive immunity based on the kinetics of responses following primary exposures to infectious agents, but their specific elements are shaped further to deliver protective subset effects under different conditions of infections. Examples of innate cellular components include the natural killer (NK) cells important in early killing of virus-infected cells and for production of early cytokines during a wide range of infections. Cellular constituents of adaptive immunity include CD4 T cells that can be driven into particular lineages to deliver subset cytokines as required for protection under different conditions of infection, and CD8 T cells mediating killing of virus-infected cells and producing cytokines for defense against viral infection. In addition, innate cytokines induced after infections are key in the endogenous responses because they can promote direct antimicrobial effects and also shape development of the cellular immune responses most beneficial under the conditions of infection being encountered.Citation1
The theme of flexibility in responding to infections is carried over to cytokine signaling and function. Although these soluble mediators have well known individual basal effects on particular cells in culture, they can have pleiotropic and overlapping effects, and depending on the conditions of examination, some of these are paradoxical. Examples include members of the Janus activated kinase-signal transducer and activator of transcription (JAK-STAT) cytokine family. These factors stimulate major signaling pathways by inducing receptor oligomerization upon ligand binding to activate receptor associated JAKs (including JAK1, JAK2, JAK3 and TYK2), which in turn recruit and activate by phosphorylation the cytoplasmic-resident STATs (including STAT1, 2, 3, 4, 5A, 5B and 6) to result in their homo- or heterodimerization, translocation into the nucleus and transcription of unique sets of target genes. Although most of these cytokines have a preference for activating particular STAT molecules, the same cytokines often have the ability to conditionally activate more than one STAT.Citation2-Citation8 This flexibility suggests that conditions for selecting particular signaling pathways could provide mechanisms for accessing different consequences of JAK-STAT cytokine exposure as needed. Thorough understanding of how this is controlled to promote important subset responses in a biological context, however, has remained elusive.
New insights are resulting from the characterization of differences in basal and induced total STAT levels as well as relative concentrations of STAT mixtures. Much of this work has focused on culture studies, but a developing literature examining responses during ongoing infection is also revealing how particular STAT concentrations are presented to modify intrinsic immune cellular responsiveness to particular cytokines and how relative concentrations of STAT combinations can be dynamically regulated to change the experience of different immune cell subsets to cytokine exposure. To date, the work has primarily focused on NK and CD8 T cell responses to type 1 interferons (IFNs) but has implications for a broader range of cells and cytokines. Taken in its entirety, it begins to explain the how and why for the complexity of reported JAK-STAT pathways as well as the pleiotropic and overlapping cytokine effects. The picture emerging is one of cytokine receptor preferences for particular STATs, but controlled variations in relative STAT concentrations to alter the selection of signaling partners.
Characterized JAK-STAT Cytokine Signaling Pathways and Functions
The original studies investigating cytokine-STAT selection used biochemical analyses to identify the major pathways delivering signals from cytokines to cells. In the case of IFNs, including type 1 (IFNα/β) and type 2 (IFNγ) IFNs, the pathways promoting their known potent antiviral effects were characterized. Type 1 IFNs are a major class of innate cytokines, comprised of products of a single IFNβ and, depending on the species, 12 IFNα genes. The type 1 IFNs activate STAT1 and STAT2, inducing their subsequent heterodimerization, and in association with the interferon regulatory factor 9 (IRF9), bind to particular DNA sequences in the promoters of genes termed IFN stimulated response elements (ISREs).Citation9 By contrast, the sole type 2 IFN, IFNγ, activates STAT1 homodimers and induces the expression of genes with gamma activating sequences (GAS) in their promoters.Citation9 The recently identified type 3 IFNs, IFNλs, also named interleukin (IL)-28A, -28B and -29, are another class of cytokines activating STAT1 and STAT2 for signaling.Citation10 Further well-characterized cytokine-STAT pathways include the activation of STAT3 by IL-6 and IL-10, the activation of STAT4 by IL-12 and the activation of STAT5 by cytokines that signal through the common gamma chain, including IL-2, IL-4, IL-7, IL-9, IL-15 and IL-21.Citation8,Citation11-Citation13
Although specific major signaling pathways are associated with exposure to particular cytokines, the factors demonstrate “infidelity” such that additional and/or other, non-preferred STATs are conditionally activated. In the most extreme case of promiscuity, the type 1 IFNs have been reported to activate all seven of the different STAT molecules (),Citation2-Citation7 but there is also a surprisingly long list of cytokines reported to activate both STAT1 and STAT3, including IFNγ, the IFNλs, IL-6, IL-10 and IL-21.Citation12,Citation14-Citation18 Because the activated STATs themselves are binding to particular promoter sequences in genes to regulate transcription, activation of common STATs by different cytokines can elicit common expression of particular target genes (). Remarkably, the type 1 IFNs can differentially regulate the expression of over 100 target genes.Citation19,Citation20 The role for STAT1 in induction of many of these, including critical antiviral genes coding for the 2,5-oligoadenylate synthetase (OAS), protein kinase R (PKR) and the Mx protein products, has been definitively established. In addition, STAT1 plays a role in the anti-proliferative and pro-apoptotic effects delivered by type 1 or type 2 IFNs,Citation21,Citation22 and the pathway contributes to certain immunoregulatory effects mediated by type 1 IFN during the development of innate immune responses to viral infections including the activation of NK cell-mediated cytotoxicity, delivered through perforin- and granzyme-dependent mechanisms, the induction of IL-15 expression and interestingly, the inhibition of IFNγ expression.Citation23-Citation25 Certain of the type 1 IFN responses can also be induced by IFNγ because it activates STAT1.Citation1,Citation26 In responses elicited by IL-12, the induction of IFNγ and the CD25 component of the IL-2 receptor is STAT4 dependent,Citation24,Citation27-Citation30 and under certain conditions, type 1 IFNs, but not IFNγ, can activate STAT4 to induce these responses (see below).Citation4,Citation31 Pro-survival states and the expression of target genes promoting these, including Bcl-XL, survivin, c-myc and cyclin D1, are linked to STAT3,Citation32-Citation35 and activation of STAT3 by the variety of cytokines might be predicted to lead to these responses. Thus, stimulation of common STATs helps explain pleiotropic and overlapping cytokine effects.
Figure 1. Flexible use of STATs by type 1 IFNs in different cells. (A) Type 1 IFNs have been shown to be able to conditionally activate all of the STATs. The key pathways, however, use the preferred STAT1/STAT2 molecules to stimulate genes through ISRE and GAS promoters for induction of an antiviral state. (B) NK cells intrinsically express high levels of STAT4. As a result, exposure to type 1 IFNs initially activates STAT4 for IFNγ production by these cells. Eventually, STAT1 levels are increased and block receptor access to STAT4. (C) CD8 T cells have the potential to respond to type 1 IFN with either STAT1 or STAT4 activation, but the activation of STAT1 is preferred. During the context of responses to infection, the antigen-specific subsets have their STAT4, whereas the non-specific cells have STAT1, levels induced. As a result, antigen-specific CD8 T cells overcome type 1 IFN STAT1-dependent anti-proliferative effects and respond to type 1 IFN with STAT4 activation for IFNγ production. (See text for related references.)
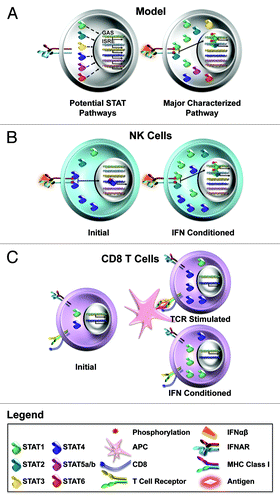
Table 1. Key cytokine pathways to STAT1, STAT3 and STAT4 functions
Flexibility in signaling may also help explain the paradoxical biological functions that have been attributed to the cytokines. Studies using cells or mice rendered genetically deficient in particular STATs have revealed that there are molecular and biological consequences of experimentally restricting access to STATs. In the absence of STAT1, the STAT1-dependent antiviral and anti-proliferative effectsCitation36-Citation38 as well as activation of NK cell-mediated cytotoxicity and induction of IL-15 expressionCitation24 are blocked during viral infections eliciting high levels of type 1 IFNs, but type 1 IFN induction of IFNγ is revealed.Citation25 Likewise, absence of STAT1 blocks the STAT1-dependent, but allows stimulation of STAT1-independent, and novel, IFNγ gene targets.Citation39 For IL-6, in the absence of STAT3, the cytokine stimulates STAT1 phosphorylation and a gene transcription program with induction of an antiviral state that resembles that induced by IFNγ.Citation40 Thus, if there are biologically relevant conditions modifying availability of particular STAT molecules, alternative signaling pathways may provide mechanisms for shaping the unique, pleiotropic, overlapping and paradoxical effects delivered by different members of the JAK-STAT cytokine family to provide pathways to particular subset responses as needed during infections. As an example, IL-10 with its potential to activate both STAT3 and STAT1 might also be driven to deliver antiviral states in the absence of STAT3. In addition to direct links between cytokine exposure and STAT activation for the induction of gene expression and rapid cellular responses discussed here, the combinations of particular cytokines with particular STATs has been shown to play a role in the differentiation of CD4 T cells into specific lineages to shape the availability of CD4 T helper cytokines under different conditions of infections. Simply stated, exposure to IL-12 uses STAT4 to support the differentiation of CD4 Th1 type cells producing IFNγ important during infections with intracellular microorganisms including certain bacteria, whereas exposure to IL-4 uses STAT6 to support the development of Th2 cells producing IL-4 important during parasitic infections, and exposure to IL-6 uses STAT3 to promote Th17 cells producing IL-17 important during infections with extracellular microorganisms including fungi.Citation41 The mechanisms involved in driving CD4 T cell subset development require longer periods and additional genetic modifications to develop.Citation42
Intrinsic Differences in STAT Expression and Use
One way of regulating access to different signaling pathways would be to provide different cell populations, with common expression of cytokine receptors, with differential expression of particular STAT levels. Because STAT1 and STAT2 are the preferred type 1 IFN receptor (IFNAR) signaling molecules, higher level expression of alternative STATs would likely be required to make them competitive in the context of STAT1/STAT2. Biochemical and flow cytometric studies evaluating STAT4 expression within freshly isolated mouse NK cells have demonstrated that these populations are unique in their basal high expression of STAT4 and modestly reduced levels of STAT1 proteins ().Citation31 This can be observed in NK cells prepared from either the spleen or the peritoneal cavity. Ex vivo type 1 IFN exposure results in the preferential activation of phosphorylated (p)STAT4 over pSTAT1 in these populations. The mouse system of lymphocytic choriomeningitis virus (LCMV) is particularly useful for studying the regulation of type 1 IFN signaling pathways in response to in vivo cytokine exposure because the virus induces a strong innate cytokine response focused on type 1 IFN production, locally detected at hours after infection and broadly observed systemically beginning from around day 1.5 and extending to day 4 of infection.Citation38,Citation43-Citation45 The very earliest type 1 IFN production at the site of infection, the peritoneal cavity, results in an IFNAR- and STAT4-dependent induction of peritoneal NK cell IFNγ expression and production of short duration, peaking at about 30 h.Citation44 This response inhibits viral replication. Thus, NK cells basally experience exposure to type 1 IFN with STAT4 rather than STAT1 activation, and this leads to IFNγ production for the benefit of an infected host.
The studies of NK cells provide the first documented differential expression of STATs in normal, freshly isolated immune populations. Other reports, however, have also shown differences in STAT4 levels in human populations isolated under disease conditionsCitation46 and in culture-derived human dendritic cell (DC) subsets.Citation47 Moreover, various populations in human peripheral blood leukocytes have different preferences for STAT activation after type 1 IFN exposure. In comparison to other cell types, exposure to the type 1 IFN, IFNβ, does not stimulate a strong pSTAT1 response but does induce pSTAT3 and pSTAT5 in B and CD4 T cells subsets.Citation48 The observation may help explain a poor induction of STAT1-dependent pro-apopotic mRNAs in human B and CD4 T cells as well as the apparent pro-survival effects of IFNβ on these cells as compared with monocytes. Although the STAT1 levels are not significantly different in the populations, however, the question of relative STAT concentrations in these different cell types has not been addressed because the levels of STAT3 and STAT5 have not been measured.Citation48 Thus, there is more work to be done with a broader range of cell subsets, and there may be other mechanisms influencing selection, but taken together, the work to date suggests a model by which differences in STAT availability based on relative concentrations influence intrinsic differential cellular responses to type 1 IFNs.
Another system with work characterizing relative access of a JAK-STAT cytokine to different STATs is the IFNγ response delivered through its receptor. STAT1 is a predominant transcription factor activated by IFNγ, but STAT3 can also be weakly activated, and in the absence of STAT1, STAT3 is highly phosphorylated and drives the expression of genes that are normally induced by IFNγ.Citation49 The sophisticated biochemical analysis of the receptor interactions with STAT1 as compared with STAT3 has demonstrated that a tyrosine residue in the IFNγ receptor subunit 1 is required for the activation of both STAT1 and STAT3, suggesting that the two STATs compete for binding to the IFNγ receptor at the same site.Citation49 Although the understanding of the STAT4/STAT1 interactions with the type 1 IFN receptor are less well developed, STAT4 is basally associated with IFNAR in the context of the high level of STAT4 and lower STAT1 within NK cells.Citation31 Thus, a growing literature indicates that the relative concentrations of different STAT molecules have consequences for signaling in response to the same cytokine. The requirements for different concentrations in STAT selection are likely to depend on the potential for, and relative affinity of, physical interactions with the respective cytokine receptors. Collectively, these studies highlight the flexibility of intrinsic STAT availability in initially selecting cytokine-driven effects and raise the interesting questions of how STAT concentrations might be regulated to influence cytokine effects during the dynamic conditions of infections to differentially shape individual cellular responses.
Kinetic Changes in STAT1 Levels: Regulating Access to Other STATs
Although type 1 IFNs can activate STAT1 and STAT4, the two STAT molecules stimulate opposing gene programs. As noted above for NK cells, activation of STAT4 promotes IFNγ production, but activation of STAT1 inhibits IFNγ expression while promoting anti-proliferative effects, cytotoxicity and IL-15 expression.Citation21,Citation24,Citation25,Citation31 The activation of STAT1 also results in increased STAT1 because the molecule targets promoter sequences in its gene to induce STAT1 mRNA and protein.Citation50,Citation51 Although this pathway has been known for some time, understanding of its biological importance remained largely unappreciated until it was demonstrated that total STAT1 levels are dramatically elevated in immune cell populations following infections of mice with LCMV.Citation4 Biochemical analysis of mixed populations has demonstrated that the ability to use type 1 IFNs for STAT4 activation inversely correlates with total STAT1 levels such that the pathway is accessible prior to infection, blocked at times with increased STAT1 levels and apparent again after the STAT1 levels have declined.Citation4 The original demonstration of this set forth the model of dynamic regulation of STAT concentrations.
Further characterization of the pathway in NK cells has shown that although these populations basally express and access STAT4 in response to type 1 IFNs, they have elevated levels of STAT1 during the course of LCMV infection.Citation31 At early times after infection, all of the immune cells examined induce STAT1 in response to type 1 IFN production, but the kinetics is delayed in NK cells relative to that of the other populations. The condition allows a window of opportunity for type 1 IFN induction of IFNγ induction though STAT4 prior to high level STAT1 expression.Citation44 The delay may be a consequence of reduced immediate activation of STAT1 because of the intrinsic lower levels of the molecule in the context of high STAT4 and/or because of competition between the high levels of STAT4 with low levels of STAT1 for access to events at the receptor.Citation44 Eventually, the STAT1 levels do increase in response to type 1 IFN exposure in the NK cells, and once elevated, they are preferentially associated with the type 1 IFN receptor such that although the STAT4 levels remain high, they are not activated by the cytokine.Citation31 The absence of STAT1 results in the ability to continue to use type 1 IFN for STAT4 activation. During LCMV infection, however, this is highly detrimental to the host because it allows for continued, dysregulated IFNγ production and cytokine-mediated disease. Thus, NK cells basally express high STAT4, and type 1 IFN induces IFNγ through this molecule to help protect against early viral replication, but STAT1 is induced to tightly regulate the pathway and protect from cytokine-mediated disease. The switch might help promote other effector functions because STAT1 is important in inducing NK cell cytotoxic function.Citation24 Thus, passing off from STAT4 to STAT1 could have many effects including changing effector functions as well as protection from immune dysregulation during infections.
Recently, NK cells and DCs isolated from another infection in mice, murine cytomegalovirus (MCMV), have been shown to exhibit different responsiveness to type 1 IFNs with alterations in STAT1 activation and induction such that the STAT1 pathway is preferentially used in DC subsets.Citation52 Remarkable aspects of the early immune response to this virus as compared with LCMV are that IL-12 is elicited along with type 1 IFNs, and high systemic levels of NK cell produced IFNγ are induced through and dependent upon IL-12 activation of STAT4.Citation24,Citation53 These responses peak at 36 to 40 h after infection. Because the IL-12 receptor preferentially activates STAT4, it is tempting to speculate that the biological pressure for IL-12 is to provide a pathway to STAT4 and IFNγ in cells that have been conditioned to express high STAT1 and as a result, have had their pathway from type 1 IFN to STAT4 blocked.
Type 1 IFN Access to STAT1 and STAT4 in CD8 T Cells after Viral Infection
Although adaptive T cells, particularly CD8 T cells, can have functions that overlap those of innate NK cells, they have unique challenges to their activation during viral infections. Foremost is that the subsets with T cell receptors (TCRs) specific for the antigens of particular infectious organisms are at extremely low frequencies in naïve, non-immune hosts and have to be selected and preferentially expanded. In the case of CD8 T cells, this occurs as a result of antigen presentation by the class 1 major histocompatibility molecules (MHC) to the TCR, takes time to develop, and during viral infections, requires proliferation through periods overlapping with the induction of systemic type 1 IFN levels and the opportunity for their delivery of anti-proliferative effects. In addition, the cells have to be driven into states that allow their needed subset responses, such as IFNγ production, and type 1 IFNs can have both inhibiting and enhancing effects on these.Citation25 The picture emerging is one where the concentrations of STAT1 and STAT4 are being differentially regulated to condition for particular experiences following exposure to type 1 IFN and as a result, facilitate the development of antigen-specific CD8 T cell responses to viral infections.
The STAT1 molecules expectedly play an important role in type 1 IFN-mediated inhibition of proliferation and have a novel role in helping to preferentially limit expansion of non-specific CD8 T cells during endogenous immune responses. In the absence of STAT1, a type 1 IFN-mediated inhibition of ex vivo cytokine-driven proliferation of CD8 T cells is lost, and LCMV infection of STAT1-deficient mice results in dysregulated early proliferation of CD8 T cells, through day 4 of infection, without specificity for the known LCMV antigens.Citation38 How then can antigen-specific cells avoid this inhibition to expand and contribute to defense? Although STAT1 is elevated in all of the splenic leukocytes isolated from mice at different times after LCMV infection, the CD8 T cells responding with DNA synthesis and expansion, on days 5 through 8 of infection, are preferentially found within subsets of cells having lower STAT1Citation38 and higher STAT4 protein levels,Citation54 and the cells expressing receptors specific for LCMV are in this group. Consistent with early reports characterizing T cell linesCitation55 and CD4 T cell subsets,Citation56 ex vivo stimulation through the TCR induces elevated STAT4 expression in CD8 T cells.Citation54 More importantly, the condition of higher STAT4 protects against type 1 IFN induction of STAT1 and resistance to type 1 IFN-mediated inhibition of proliferation ex vivo, and the presence of STAT4 results in enhanced proliferation and reduced STAT1 expression in antigen-specific CD8 T cells during LCMV infection.Citation54 Thus, CD8 T cell subsets are being differentially conditioned () such that the non-specific cells have STAT1 levels induced and are sensitive to type 1 IFN-mediated inhibition of proliferation, and the antigen-specific cells are induced to express higher levels of STAT4 and as a result, develop resistance to type 1 IFN-mediated inhibition of proliferation with selection for preferential expansion in the presence of the cytokines.
The changing STAT concentrations also have consequences for cellular effector responses to viral infections. Although the numbers of antigen-specific CD8 T cells are still difficult to evaluate at days 4 to 6 of LCMV infection, a systemic CD8 T cell-dependent IFNγ response is detectable at these times.Citation45 In vivo blocking studies have shown that the response is primarily dependent on antigen-specific CD8 T cells and is greatly enhanced through type 1 IFN- and STAT4-dependent pathways.Citation4,Citation45 Dissection of the components of stimulation ex vivo shows that there is a high degree of synergism such that although either works alone, small concentrations of type 1 IFN along with small concentrations of antigen enhance IFNγ production.Citation45 Type 1 IFN responses of CD8 T cells prepared from uninfected as compared with day 8 LCMV-infected mice, switch from preferentially activating STAT1 and a wide range of target genes, including those dependent on this transcription factor, to preferentially activating STAT4 and focusing the responses to a narrower target gene range with less expression of the STAT1 but continued or elevated expression of the STAT4 target genes.Citation54 These observations explain the differential effects of type 1 IFN on proliferation and IFNγ expression by showing that the CD8 T cells are being conditioned to experience the cytokines differentially, and that this is important in shaping the endogenous adaptive immune responses to viral infection. It is interesting to note that the antigen-specific CD8 T cells now mirror the basal STAT expression of NK cells, high for STAT4 and lower for STAT1.
STAT3: A Role in Conditioning Lymphocyte Responses to Cytokines after Viral Infections?
How might regulation of STAT3 access fit into intrinsic or conditioned cytokine responses during viral infections? STAT3 can be activated by many of the same cytokines as STAT1, but the functional consequences have been difficult to assign and as noted above, are often cell type and context dependent. The molecule was initially discovered to mediate the acute phase response in hepatocytes in response to IL-6, resulting in the transcription of genes that play a protective role (e.g., wound healing) in acute inflammation.Citation11,Citation57,Citation58 The IL-6-STAT3 pathway has also been shown to prevent apoptosis in both pro-B cell lines and primary T cells, in the former case by STAT3-mediated induction of Bcl-2, a member of the anti-apoptotic Bcl family.Citation59,Citation60 In the case of type 1 IFN effects on lymphocyte proliferation or survival, the role for STAT3 has again been controversial with reports of its positive action being context dependent.Citation61,Citation62 STAT3 is the major transducer of IL-10 signaling, and in macrophages, this pathway both limits inflammatory cytokine production and prevents proliferation.Citation63,Citation64 In STAT3-deficient macrophages, the cells produce increased levels of inflammatory cytokines in response to the bacterial product, lipopolysaccharide (LPS), resulting in chronic enterocolitis in mice.Citation65 Despite these varying functional outcomes in different cell types, an interesting feature of STAT3 is that, similar to STAT1, the protein levels of STAT3 are increased when STAT3 is activated by cytokines.Citation66,Citation67 Thus, the opportunity to condition cellular responses based on modulation of STAT3 levels represents a viable mechanism for controlling cellular functions. Moreover, given the wide range of receptors stimulating STAT3, there is a potential for variations in particular STAT levels relative to other STATs to have very different consequences for shaping the responses to particular cytokines.
Because the molecule is also associated with key cellular processes, including survival, proliferation and apoptosis, understanding the regulation of expression and access could help explain complex events preferentially supporting expansion and maintenance of needed lymphocytes. A STAT3 requirement for survival is underscored by the fact that unlike mice deficient in other STAT molecules, mice with targeted deletion of STAT3 do not develop past embryonic day 6.5.Citation68 However, conditional deletion of STAT3 in various cell types has not always presented a consistent picture of STAT3 in promoting survival. For example, deficiency of STAT3 in neurons results in enhanced apoptosis of these cells,Citation69 whereas STAT3 deletion in the mammary epithelium delays the process of involution depending on apoptosis.Citation70 Interestingly, the most striking evidence for STAT3 mediating survival signals is the multitude of studies highlighting the involvement of STAT3 in cellular transformation. The molecule is constitutively activated in many human tumors,Citation71-Citation73 and cells with genetically mutated STAT3 are resistant to transformation,Citation74 whereas cells expressing a constitutively active form of STAT3 are converted to cancer cells.Citation75 Consistent with the latter, many of the well-documented target genes of STAT3 include those that prevent apoptosis (Bcl-xl and survivin) and enhance survival and proliferation (cyclin D1 and c-myc).Citation32-Citation35 Thus, there is much to be learned about how STAT3 is used in the context of evolving immune responses to infection. Given that type 1 IFNs can mediate different NK and T cell responses based on their ability to signal through STAT1 or STAT4, and their preference is influenced by the levels of the STATs, infections of mice with LCMV inducing type 1 IFNs or MCMV inducing additional cytokines, including IFNγ and IL-6,Citation76 provide conditions for changing STAT3 levels in responding cells and give the potential for adding this signaling molecule to the mix of relative STAT concentrations shaping cellular responses to STAT3 activating cytokines. Future studies are needed to better define the modulation of interaction between these STATs in the context of a complex cytokine milieu, and studies of viral infections hold promise for evaluating the contributions of regulated STAT3 expression in shaping cellular immune functions.
Summary and Discussion
Taken together, the studies of NK and CD8 T cells in uninfected and virus-infected mice have demonstrated differential type 1 IFN responsiveness based on intrinsic and induced STAT concentrations. The STAT4 levels are basally high in NK cells, allowing them a window of opportunity to immediately respond to type 1 IFNs with IFNγ production, but the pathway is tightly regulated by simultaneous STAT1 induction to protect from cytokine-mediated disease. In contrast, antigen recognition induces STAT4 in selected CD8 T cells while STAT1 is elevated in non-specific CD8 T cell subsets to allow for preferential expansion of, and type 1 IFN induction of IFNγ in, the antigen-specific subsets. The flexible pathways to STAT4 allow general access to a highly controlled early NK cell response followed by access to an antigen-specific CD8 T cell response. Because the CD8 T cell induction of IFNγ is greatly enhanced by TCR stimulation as well as type 1 IFN, the conditions limit this response to periods overlapping viral replication for antigen presentation and type 1 IFN production (). Thus, their STAT4-dependent type 1 IFN responses are controlled even though STAT1 is not elevated to high levels. The sophisticated kinetic regulation of these events provides insights into how the immune system uses the resources available to promote optimal defense against infection while protecting against potential immune-mediated disease. They also suggest approaches for therapeutic intervention by identifying the when and how particular pathways are required.
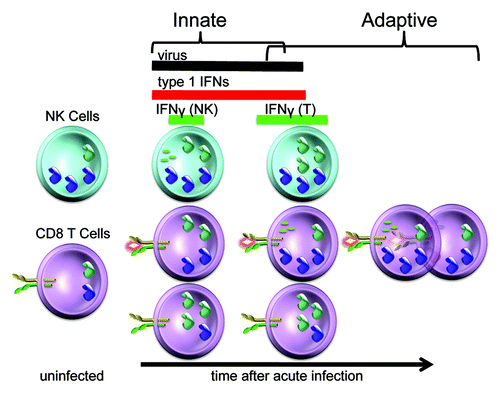
Figure 2. Differential cellular use of type 1 IFNs for STAT4 activation and IFNγ production at particular times during LCMV infection. The LCMV infection in mice provides a powerful system to study the regulation of type 1 IFN effects. The cytokines are produced locally within a few hours and can be found systemically for several days following infection. Prior to infection, NK cell populations are high whereas the CD8 T cells are low for STAT4. As a result, NK cells initially respond to type 1 IFN with STAT4 activation and IFNγ production. The pathway is tightly regulated, however, because elevated STAT1 levels are concurrently induced to block type 1 IFN access to STAT4. In contrast, the antigen-specific CD8 T cells are being stimulated through their TCR to have elevated STAT4 expression whereas the non-specific CD8 T cells are induced to express elevated STAT1. These conditions promote preferential expansion of the antigen-specific CD8 T cells and their production of IFNγ in the context of the endogenous type 1 IFNs. Thus, there are flexible pathways in different cell types to regulate the consequences of type 1 IFN exposure during viral infection. (See text for related references.)
Although most of the work reviewed here has been performed in the mouse, parallel systems operate in the human. Indeed, the first report of type 1 IFN activation of STAT4 examined responses in human T cells,Citation77 and more recent work has shown the response in human NK cells.Citation78 In fact, conflicting data suggesting that there were species differences in type 1 IFN activation of STAT4 were resolved by the demonstration of differential activation of STAT4 based on STAT1 concentrations in the mouse,Citation4 and the correlation between type 1 IFN activation of STAT4 with low STAT1 levels has been recently reported with cells from individuals chronically infected with hepatitis C virus (HCV) as compared with control populations.Citation78,Citation79 Thus, the work in the human and mouse is crystallizing to show how partners at the dance can be changed based on availability and concentrations to alter STAT pathways of activation and cytokine functions under particular conditions of infection.
There are still many unanswered questions. What other events control differential intrinsic STAT concentrations? Does constitutive type 1 IFN signaling help set intrinsic STAT levels in particular cells?Citation80 Why is the block in STAT1 induction in antigen-specific CD8 T cells more complete than the delayed induction in NK cells even though both cell types have elevated STAT4? The results comparing the LCMV and MCMV infection systems suggest that IL-12 access to STAT4 is relatively insensitive to high STAT1 levels, but almost nothing else is known about how the altered STAT4 and STAT1 concentrations might affect cellular responses to the range of additional, non-type 1 IFN cytokines having flexibility to stimulate these molecules. Moreover, there are a variety of pathways for regulating STAT concentrations. The pathways for STAT1 activation to elevated STAT1 expression and from TCR stimulation to elevated STAT4 have been shown to work during precise periods of viral infections (). There are also pathways from STAT3 activation to elevated STAT3 expression, however, and many other yet unidentified mechanisms are likely to be available to regulate relative concentrations of all of the STATs in mixtures (). Characterization of these, how they function during immune responses and how they change signaling of the range of cytokines with the potential to activate them will provide novel new insights into how immune responses are regulated.
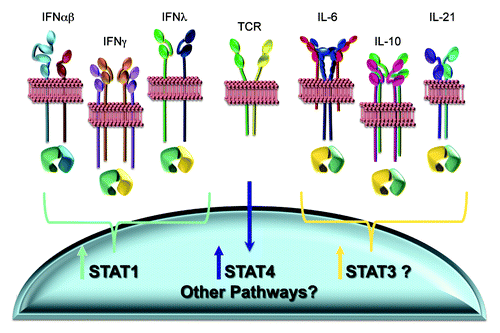
Figure 3. Known and potential pathways for changing intracellular STAT concentrations. A variety of cytokines, including all of the IFNs, preferentially activate STAT1, and STAT1 activation results in the induction of elevated STAT1 expression. Stimulation through the TCR leads to elevated STAT4. These pathways have been shown to be operational during viral infections. Other cytokines, including IL-6, IL-10 and IL-21, can activate STAT1 but have a preference for activating STAT3. STAT3 is reported to induce its own expression. Certain of these factors are also elicited during viral infections. Thus, there are a variety of potential known mechanisms for altering STAT3, and the potential for many unknown mechanisms for altering additional STATs, to further change intracellular concentrations of STAT mixes and the effects of any cytokine with the ability to alternatively activate particular STATs. (See text for related references.)
The results demonstrating the dynamic changes in particular STAT concentrations suggest that care must be taken in interpreting long-term studies of the role for particular cytokinesCitation81 or STATsCitation82 after transfer of deficient cell subsets into complete environments. Clearly, both the cytokines and the STATs are differentially used during the life of particular cells. Thus, although such long-term studies may provide information on “net” outcomes, they do not present opportunities for identifying the regulation of access to STATs for changing cell or host need, or for revealing key functions depending on particular STATs or cytokines that might be delivered downstream of the blocked pathway being tested. The same caution can be raised about the powerful approach of identifying genetic susceptibilities to classes of infectious organisms to assign unique importance of particular receptors and STATs in defense.Citation83-Citation91 The outcome does reveal absolute requirements but does not inform on other effects, flexible pathways and/or on alternative pathways providing other kinds of support. Indeed, in the case of STAT3 mutations in humans, the first reports linked the condition to increased susceptibility to fungal infections.Citation88,Citation89 The consequences for long-term maintenance of T cell subsets and control of viral infections were only revealed after more extensive and detailed studies.Citation92 Thus, a thorough understanding of the mechanisms shaping cytokine and STAT functions requires careful dissection of cellular responses in the context of the evolving conditions of different periods of infections.
In conclusion, there is still much to be learned about regulation of cytokine effects and signaling pathways, but existing evidence provides a strong model for continued testing. Clearly, biological outcomes will depend on the integration of multiple rather than additive effects of individual STAT signals. However, the results to date indicate that these are subject to conditioning based on the dynamic regulation of relative STAT concentrations. The consequences of varying STAT levels can be predicted to depend on the potential for, and relative affinity of, physical interactions with the respective cytokine receptors. Given the range of cytokines activating overlapping STATs, the opportunities for differential regulation of their effects with changing STAT concentrations are great. With the framework of existing knowledge, the next period of study will be exciting and lead to new understanding about how the immune response is working in its entirety to achieve the best possible response to infections using the available cytokine and signaling resources. Future work holds great promise for not only explaining the how and why endogenous immune responses unfold the way they do, but also for providing insights into new approaches for intervention to promote health over disease in a variety of challenging conditions.
| Abbreviations: | ||
| NK | = | natural killer cells |
| LCMV | = | lymphocytic choriomeningitis virus |
| MCMV | = | murine cytomegalovirus |
| HCV | = | hepatitis C virus |
Acknowledgments
Work from the Biron laboratory has been supported by National Institutes of Health USA grants CA41268 and AI55677. The authors thank their current and previous laboratory members and collaborators for the work leading to the concepts presented here, particularly Ken Nguyen, Pilar Gil, Takuya Miyagi, Mickaël Ploquin, Ethan Mack, Delia Demers, Wendy Watford, Yuka Kanno and John O’Shea as well as Mimi Fragoso for help with the figures. They apologize for the work of colleagues that might not have been included due to space restraints.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- Biron CA, Sen G. C. In: Knipe DMea, ed. Fields Virology, Fourth Edition: Lippincott, Williams, and Wilkins, 2001.
- Brierley MM, Fish EN. Review: IFN-alpha/beta receptor interactions to biologic outcomes: understanding the circuitry. J Interferon Cytokine Res 2002; 22:835 - 45; http://dx.doi.org/10.1089/107999002760274845; PMID: 12396722
- Cho SS, Bacon CM, Sudarshan C, Rees RC, Finbloom D, Pine R, et al. Activation of STAT4 by IL-12 and IFN-alpha: evidence for the involvement of ligand-induced tyrosine and serine phosphorylation. J Immunol 1996; 157:4781 - 9; PMID: 8943379
- Nguyen KB, Watford WT, Salomon R, Hofmann SR, Pien GC, Morinobu A, et al. Critical role for STAT4 activation by type 1 interferons in the interferon-gamma response to viral infection. Science 2002; 297:2063 - 6; http://dx.doi.org/10.1126/science.1074900; PMID: 12242445
- van Boxel-Dezaire AH, Rani MR, Stark GR. Complex modulation of cell type-specific signaling in response to type I interferons. Immunity 2006; 25:361 - 72; http://dx.doi.org/10.1016/j.immuni.2006.08.014; PMID: 16979568
- Ho HH, Ivashkiv LB. Role of STAT3 in type I interferon responses. Negative regulation of STAT1-dependent inflammatory gene activation. J Biol Chem 2006; 281:14111 - 8; http://dx.doi.org/10.1074/jbc.M511797200; PMID: 16571725
- García-Sastre A, Biron CA. Type 1 interferons and the virus-host relationship: a lesson in détente. Science 2006; 312:879 - 82; http://dx.doi.org/10.1126/science.1125676; PMID: 16690858
- Rochman Y, Spolski R, Leonard WJ. New insights into the regulation of T cells by gamma(c) family cytokines. Nat Rev Immunol 2009; 9:480 - 90; http://dx.doi.org/10.1038/nri2580; PMID: 19543225
- Darnell JE Jr., Kerr IM, Stark GR. Jak-STAT pathways and transcriptional activation in response to IFNs and other extracellular signaling proteins. Science 1994; 264:1415 - 21; http://dx.doi.org/10.1126/science.8197455; PMID: 8197455
- Kotenko SV, Gallagher G, Baurin VV, Lewis-Antes A, Shen M, Shah NK, et al. IFN-lambdas mediate antiviral protection through a distinct class II cytokine receptor complex. Nat Immunol 2003; 4:69 - 77; http://dx.doi.org/10.1038/ni875; PMID: 12483210
- Zhong Z, Wen Z, Darnell JE Jr.. Stat3: a STAT family member activated by tyrosine phosphorylation in response to epidermal growth factor and interleukin-6. Science 1994; 264:95 - 8; http://dx.doi.org/10.1126/science.8140422; PMID: 8140422
- Lai CF, Ripperger J, Morella KK, Jurlander J, Hawley TS, Carson WE, et al. Receptors for interleukin (IL)-10 and IL-6-type cytokines use similar signaling mechanisms for inducing transcription through IL-6 response elements. J Biol Chem 1996; 271:13968 - 75; http://dx.doi.org/10.1074/jbc.271.24.13968; PMID: 8662928
- Jacobson NG, Szabo SJ, Weber-Nordt RM, Zhong Z, Schreiber RD, Darnell JE Jr., et al. Interleukin 12 signaling in T helper type 1 (Th1) cells involves tyrosine phosphorylation of signal transducer and activator of transcription (Stat)3 and Stat4. J Exp Med 1995; 181:1755 - 62; http://dx.doi.org/10.1084/jem.181.5.1755; PMID: 7722452
- Caldenhoven E, Buitenhuis M, van Dijk TB, Raaijmakers JA, Lammers JW, Koenderman L, et al. Lineage-specific activation of STAT3 by interferon-gamma in human neutrophils. J Leukoc Biol 1999; 65:391 - 6; PMID: 10080544
- Shuai K, Schindler C, Prezioso VR, Darnell JE Jr.. Activation of transcription by IFN-gamma: tyrosine phosphorylation of a 91-kD DNA binding protein. Science 1992; 258:1808 - 12; http://dx.doi.org/10.1126/science.1281555; PMID: 1281555
- Donnelly RP, Sheikh F, Kotenko SV, Dickensheets H. The expanded family of class II cytokines that share the IL-10 receptor-2 (IL-10R2) chain. J Leukoc Biol 2004; 76:314 - 21; http://dx.doi.org/10.1189/jlb.0204117; PMID: 15123776
- Finbloom DS, Winestock KD. IL-10 induces the tyrosine phosphorylation of tyk2 and Jak1 and the differential assembly of STAT1 alpha and STAT3 complexes in human T cells and monocytes. J Immunol 1995; 155:1079 - 90; PMID: 7543512
- Habib T, Nelson A, Kaushansky K. IL-21: a novel IL-2-family lymphokine that modulates B, T, and natural killer cell responses. J Allergy Clin Immunol 2003; 112:1033 - 45; http://dx.doi.org/10.1016/j.jaci.2003.08.039; PMID: 14657853
- Der SD, Zhou A, Williams BR, Silverman RH. Identification of genes differentially regulated by interferon alpha, beta, or gamma using oligonucleotide arrays. Proc Natl Acad Sci U S A 1998; 95:15623 - 8; http://dx.doi.org/10.1073/pnas.95.26.15623; PMID: 9861020
- Schoggins JW, Wilson SJ, Panis M, Murphy MY, Jones CT, Bieniasz P, et al. A diverse range of gene products are effectors of the type I interferon antiviral response. Nature 2011; 472:481 - 5; http://dx.doi.org/10.1038/nature09907; PMID: 21478870
- Bromberg JF, Horvath CM, Wen Z, Schreiber RD, Darnell JE Jr.. Transcriptionally active Stat1 is required for the antiproliferative effects of both interferon alpha and interferon gamma. Proc Natl Acad Sci U S A 1996; 93:7673 - 8; http://dx.doi.org/10.1073/pnas.93.15.7673; PMID: 8755534
- Ramana CV, Grammatikakis N, Chernov M, Nguyen H, Goh KC, Williams BR, et al. Regulation of c-myc expression by IFN-gamma through Stat1-dependent and -independent pathways. EMBO J 2000; 19:263 - 72; http://dx.doi.org/10.1093/emboj/19.2.263; PMID: 10637230
- Lee CK, Rao DT, Gertner R, Gimeno R, Frey AB, Levy DE. Distinct requirements for IFNs and STAT1 in NK cell function. J Immunol 2000; 165:3571 - 7; PMID: 11034357
- Nguyen KB, Salazar-Mather TP, Dalod MY, Van Deusen JB, Wei XQ, Liew FY, et al. Coordinated and distinct roles for IFN-alpha beta, IL-12, and IL-15 regulation of NK cell responses to viral infection. J Immunol 2002; 169:4279 - 87; PMID: 12370359
- Nguyen KB, Cousens LP, Doughty LA, Pien GC, Durbin JE, Biron CA. Interferon alpha/beta-mediated inhibition and promotion of interferon gamma: STAT1 resolves a paradox. Nat Immunol 2000; 1:70 - 6; http://dx.doi.org/10.1038/76940; PMID: 10881178
- Shuai K, Stark GR, Kerr IM, Darnell JE Jr.. A single phosphotyrosine residue of Stat91 required for gene activation by interferon-gamma. Science 1993; 261:1744 - 6; http://dx.doi.org/10.1126/science.7690989; PMID: 7690989
- Barbulescu K, Becker C, Schlaak JF, Schmitt E, Meyer zum Büschenfelde KH, Neurath MF. IL-12 and IL-18 differentially regulate the transcriptional activity of the human IFN-gamma promoter in primary CD4+ T lymphocytes. J Immunol 1998; 160:3642 - 7; PMID: 9558063
- Lawless VA, Zhang S, Ozes ON, Bruns HA, Oldham I, Hoey T, et al. Stat4 regulates multiple components of IFN-gamma-inducing signaling pathways. J Immunol 2000; 165:6803 - 8; PMID: 11120802
- O’Sullivan A, Chang HC, Yu Q, Kaplan MH. STAT4 is required for interleukin-12-induced chromatin remodeling of the CD25 locus. J Biol Chem 2004; 279:7339 - 45; http://dx.doi.org/10.1074/jbc.M309979200; PMID: 14660657
- Lee SH, Fragoso MF, Biron CA. Cutting edge: a novel mechanism bridging innate and adaptive immunity: IL-12 induction of CD25 to form high-affinity IL-2 receptors on NK cells. J Immunol 2012; 189:2712 - 6; http://dx.doi.org/10.4049/jimmunol.1201528; PMID: 22888135
- Miyagi T, Gil MP, Wang X, Louten J, Chu WM, Biron CA. High basal STAT4 balanced by STAT1 induction to control type 1 interferon effects in natural killer cells. J Exp Med 2007; 204:2383 - 96; http://dx.doi.org/10.1084/jem.20070401; PMID: 17846149
- Catlett-Falcone R, Landowski TH, Oshiro MM, Turkson J, Levitzki A, Savino R, et al. Constitutive activation of Stat3 signaling confers resistance to apoptosis in human U266 myeloma cells. Immunity 1999; 10:105 - 15; http://dx.doi.org/10.1016/S1074-7613(00)80011-4; PMID: 10023775
- Gritsko T, Williams A, Turkson J, Kaneko S, Bowman T, Huang M, et al. Persistent activation of stat3 signaling induces survivin gene expression and confers resistance to apoptosis in human breast cancer cells. Clin Cancer Res 2006; 12:11 - 9; http://dx.doi.org/10.1158/1078-0432.CCR-04-1752; PMID: 16397018
- Leslie K, Lang C, Devgan G, Azare J, Berishaj M, Gerald W, et al. Cyclin D1 is transcriptionally regulated by and required for transformation by activated signal transducer and activator of transcription 3. Cancer Res 2006; 66:2544 - 52; http://dx.doi.org/10.1158/0008-5472.CAN-05-2203; PMID: 16510571
- Kiuchi N, Nakajima K, Ichiba M, Fukada T, Narimatsu M, Mizuno K, et al. STAT3 is required for the gp130-mediated full activation of the c-myc gene. J Exp Med 1999; 189:63 - 73; http://dx.doi.org/10.1084/jem.189.1.63; PMID: 9874564
- Meraz MA, White JM, Sheehan KC, Bach EA, Rodig SJ, Dighe AS, et al. Targeted disruption of the Stat1 gene in mice reveals unexpected physiologic specificity in the JAK-STAT signaling pathway. Cell 1996; 84:431 - 42; http://dx.doi.org/10.1016/S0092-8674(00)81288-X; PMID: 8608597
- Durbin JE, Hackenmiller R, Simon MC, Levy DE. Targeted disruption of the mouse Stat1 gene results in compromised innate immunity to viral disease. Cell 1996; 84:443 - 50; http://dx.doi.org/10.1016/S0092-8674(00)81289-1; PMID: 8608598
- Gil MP, Salomon R, Louten J, Biron CA. Modulation of STAT1 protein levels: a mechanism shaping CD8 T-cell responses in vivo. Blood 2006; 107:987 - 93; http://dx.doi.org/10.1182/blood-2005-07-2834; PMID: 16210337
- Gil MP, Bohn E, O’Guin AK, Ramana CV, Levine B, Stark GR, et al. Biologic consequences of Stat1-independent IFN signaling. Proc Natl Acad Sci U S A 2001; 98:6680 - 5; http://dx.doi.org/10.1073/pnas.111163898; PMID: 11390995
- Costa-Pereira AP, Tininini S, Strobl B, Alonzi T, Schlaak JF, Is’harc H, et al. Mutational switch of an IL-6 response to an interferon-gamma-like response. Proc Natl Acad Sci U S A 2002; 99:8043 - 7; http://dx.doi.org/10.1073/pnas.122236099; PMID: 12060750
- Zhu J, Yamane H, Paul WE. Differentiation of effector CD4 T cell populations (*). Annu Rev Immunol 2010; 28:445 - 89; http://dx.doi.org/10.1146/annurev-immunol-030409-101212; PMID: 20192806
- Kanno Y, Vahedi G, Hirahara K, Singleton K, O’Shea JJ. Transcriptional and epigenetic control of T helper cell specification: molecular mechanisms underlying commitment and plasticity. Annu Rev Immunol 2012; 30:707 - 31; http://dx.doi.org/10.1146/annurev-immunol-020711-075058; PMID: 22224760
- Biron CA, Nguyen KB, Pien GC. Innate immune responses to LCMV infections: natural killer cells and cytokines. Curr Top Microbiol Immunol 2002; 263:7 - 27; http://dx.doi.org/10.1007/978-3-642-56055-2_2; PMID: 11987821
- Mack EA, Kallal LE, Demers DA, Biron CA. Type 1 interferon induction of natural killer cell gamma interferon production for defense during lymphocytic choriomeningitis virus infection. MBio 2011; 2; http://dx.doi.org/10.1128/mBio.00169-11; PMID: 21828218
- Pien GC, Nguyen KB, Malmgaard L, Satoskar AR, Biron CA. A unique mechanism for innate cytokine promotion of T cell responses to viral infections. J Immunol 2002; 169:5827 - 37; PMID: 12421964
- Frucht DM, Aringer M, Galon J, Danning C, Brown M, Fan S, et al. Stat4 is expressed in activated peripheral blood monocytes, dendritic cells, and macrophages at sites of Th1-mediated inflammation. J Immunol 2000; 164:4659 - 64; PMID: 10779770
- Longman RS, Braun D, Pellegrini S, Rice CM, Darnell RB, Albert ML. Dendritic-cell maturation alters intracellular signaling networks, enabling differential effects of IFN-alpha/beta on antigen cross-presentation. Blood 2007; 109:1113 - 22; http://dx.doi.org/10.1182/blood-2006-05-023465; PMID: 17018853
- van Boxel-Dezaire AH, Zula JA, Xu Y, Ransohoff RM, Jacobberger JW, Stark GR. Major differences in the responses of primary human leukocyte subsets to IFN-beta. J Immunol 2010; 185:5888 - 99; http://dx.doi.org/10.4049/jimmunol.0902314; PMID: 20956346
- Qing Y, Stark GR. Alternative activation of STAT1 and STAT3 in response to interferon-gamma. J Biol Chem 2004; 279:41679 - 85; http://dx.doi.org/10.1074/jbc.M406413200; PMID: 15284232
- Lehtonen A, Matikainen S, Julkunen I. Interferons up-regulate STAT1, STAT2, and IRF family transcription factor gene expression in human peripheral blood mononuclear cells and macrophages. J Immunol 1997; 159:794 - 803; PMID: 9218597
- Wong LH, Sim H, Chatterjee-Kishore M, Hatzinisiriou I, Devenish RJ, Stark G, et al. Isolation and characterization of a human STAT1 gene regulatory element. Inducibility by interferon (IFN) types I and II and role of IFN regulatory factor-1. J Biol Chem 2002; 277:19408 - 17; http://dx.doi.org/10.1074/jbc.M111302200; PMID: 11909852
- Baranek T, Manh TP, Alexandre Y, Maqbool MA, Cabeza JZ, Tomasello E, et al. Differential responses of immune cells to type I interferon contribute to host resistance to viral infection. Cell Host Microbe 2012; 12:571 - 84; http://dx.doi.org/10.1016/j.chom.2012.09.002; PMID: 23084923
- Orange JS, Biron CA. An absolute and restricted requirement for IL-12 in natural killer cell IFN-gamma production and antiviral defense. Studies of natural killer and T cell responses in contrasting viral infections. J Immunol 1996; 156:1138 - 42; PMID: 8557990
- Gil MP, Ploquin MJ, Watford WT, Lee SH, Kim K, Wang X, et al. Regulating type 1 IFN effects in CD8 T cells during viral infections: changing STAT4 and STAT1 expression for function. Blood 2012; 120:3718 - 28; http://dx.doi.org/10.1182/blood-2012-05-428672; PMID: 22968462
- Bacon CM, Petricoin EF 3rd, Ortaldo JR, Rees RC, Larner AC, Johnston JA, et al. Interleukin 12 induces tyrosine phosphorylation and activation of STAT4 in human lymphocytes. Proc Natl Acad Sci U S A 1995; 92:7307 - 11; http://dx.doi.org/10.1073/pnas.92.16.7307; PMID: 7638186
- Watford WT, Hissong BD, Durant LR, Yamane H, Muul LM, Kanno Y, et al. Tpl2 kinase regulates T cell interferon-gamma production and host resistance to Toxoplasma gondii. J Exp Med 2008; 205:2803 - 12; http://dx.doi.org/10.1084/jem.20081461; PMID: 19001140
- Wegenka UM, Buschmann J, Lütticken C, Heinrich PC, Horn F. Acute-phase response factor, a nuclear factor binding to acute-phase response elements, is rapidly activated by interleukin-6 at the posttranslational level. Mol Cell Biol 1993; 13:276 - 88; PMID: 7678052
- Akira S, Nishio Y, Inoue M, Wang XJ, Wei S, Matsusaka T, et al. Molecular cloning of APRF, a novel IFN-stimulated gene factor 3 p91-related transcription factor involved in the gp130-mediated signaling pathway. Cell 1994; 77:63 - 71; http://dx.doi.org/10.1016/0092-8674(94)90235-6; PMID: 7512451
- Fukada T, Hibi M, Yamanaka Y, Takahashi-Tezuka M, Fujitani Y, Yamaguchi T, et al. Two signals are necessary for cell proliferation induced by a cytokine receptor gp130: involvement of STAT3 in anti-apoptosis. Immunity 1996; 5:449 - 60; http://dx.doi.org/10.1016/S1074-7613(00)80501-4; PMID: 8934572
- Takeda K, Kaisho T, Yoshida N, Takeda J, Kishimoto T, Akira S. Stat3 activation is responsible for IL-6-dependent T cell proliferation through preventing apoptosis: generation and characterization of T cell-specific Stat3-deficient mice. J Immunol 1998; 161:4652 - 60; PMID: 9794394
- Gimeno R, Lee CK, Schindler C, Levy DE. Stat1 and Stat2 but not Stat3 arbitrate contradictory growth signals elicited by alpha/beta interferon in T lymphocytes. Mol Cell Biol 2005; 25:5456 - 65; http://dx.doi.org/10.1128/MCB.25.13.5456-5465.2005; PMID: 15964802
- Tanabe Y, Nishibori T, Su L, Arduini RM, Baker DP, David M. Cutting edge: role of STAT1, STAT3, and STAT5 in IFN-alpha beta responses in T lymphocytes. J Immunol 2005; 174:609 - 13; PMID: 15634877
- O’Farrell AM, Liu Y, Moore KW, Mui AL. IL-10 inhibits macrophage activation and proliferation by distinct signaling mechanisms: evidence for Stat3-dependent and -independent pathways. EMBO J 1998; 17:1006 - 18; http://dx.doi.org/10.1093/emboj/17.4.1006; PMID: 9463379
- Riley JK, Takeda K, Akira S, Schreiber RD. Interleukin-10 receptor signaling through the JAK-STAT pathway. Requirement for two distinct receptor-derived signals for anti-inflammatory action. J Biol Chem 1999; 274:16513 - 21; http://dx.doi.org/10.1074/jbc.274.23.16513; PMID: 10347215
- Takeda K, Clausen BE, Kaisho T, Tsujimura T, Terada N, Förster I, et al. Enhanced Th1 activity and development of chronic enterocolitis in mice devoid of Stat3 in macrophages and neutrophils. Immunity 1999; 10:39 - 49; http://dx.doi.org/10.1016/S1074-7613(00)80005-9; PMID: 10023769
- Narimatsu M, Maeda H, Itoh S, Atsumi T, Ohtani T, Nishida K, et al. Tissue-specific autoregulation of the stat3 gene and its role in interleukin-6-induced survival signals in T cells. Mol Cell Biol 2001; 21:6615 - 25; http://dx.doi.org/10.1128/MCB.21.19.6615-6625.2001; PMID: 11533249
- Yang J, Chatterjee-Kishore M, Staugaitis SM, Nguyen H, Schlessinger K, Levy DE, et al. Novel roles of unphosphorylated STAT3 in oncogenesis and transcriptional regulation. Cancer Res 2005; 65:939 - 47; PMID: 15705894
- Takeda K, Noguchi K, Shi W, Tanaka T, Matsumoto M, Yoshida N, et al. Targeted disruption of the mouse Stat3 gene leads to early embryonic lethality. Proc Natl Acad Sci U S A 1997; 94:3801 - 4; http://dx.doi.org/10.1073/pnas.94.8.3801; PMID: 9108058
- Alonzi T, Middleton G, Wyatt S, Buchman V, Betz UA, Müller W, et al. Role of STAT3 and PI 3-kinase/Akt in mediating the survival actions of cytokines on sensory neurons. Mol Cell Neurosci 2001; 18:270 - 82; http://dx.doi.org/10.1006/mcne.2001.1018; PMID: 11591128
- Chapman RS, Lourenco PC, Tonner E, Flint DJ, Selbert S, Takeda K, et al. Suppression of epithelial apoptosis and delayed mammary gland involution in mice with a conditional knockout of Stat3. Genes Dev 1999; 13:2604 - 16; http://dx.doi.org/10.1101/gad.13.19.2604; PMID: 10521404
- Sartor CI, Dziubinski ML, Yu CL, Jove R, Ethier SP. Role of epidermal growth factor receptor and STAT-3 activation in autonomous proliferation of SUM-102PT human breast cancer cells. Cancer Res 1997; 57:978 - 87; PMID: 9041204
- Weber-Nordt RM, Egen C, Wehinger J, Ludwig W, Gouilleux-Gruart V, Mertelsmann R, et al. Constitutive activation of STAT proteins in primary lymphoid and myeloid leukemia cells and in Epstein-Barr virus (EBV)-related lymphoma cell lines. Blood 1996; 88:809 - 16; PMID: 8704235
- Zhang Q, Nowak I, Vonderheid EC, Rook AH, Kadin ME, Nowell PC, et al. Activation of Jak/STAT proteins involved in signal transduction pathway mediated by receptor for interleukin 2 in malignant T lymphocytes derived from cutaneous anaplastic large T-cell lymphoma and Sezary syndrome. Proc Natl Acad Sci U S A 1996; 93:9148 - 53; http://dx.doi.org/10.1073/pnas.93.17.9148; PMID: 8799169
- Chiarle R, Simmons WJ, Cai H, Dhall G, Zamo A, Raz R, et al. Stat3 is required for ALK-mediated lymphomagenesis and provides a possible therapeutic target. Nat Med 2005; 11:623 - 9; http://dx.doi.org/10.1038/nm1249; PMID: 15895073
- Bromberg JF, Wrzeszczynska MH, Devgan G, Zhao Y, Pestell RG, Albanese C, et al. Stat3 as an oncogene. Cell 1999; 98:295 - 303; http://dx.doi.org/10.1016/S0092-8674(00)81959-5; PMID: 10458605
- Ruzek MC, Miller AH, Opal SM, Pearce BD, Biron CA. Characterization of early cytokine responses and an interleukin (IL)-6-dependent pathway of endogenous glucocorticoid induction during murine cytomegalovirus infection. J Exp Med 1997; 185:1185 - 92; http://dx.doi.org/10.1084/jem.185.7.1185; PMID: 9104805
- Rogge L, D’Ambrosio D, Biffi M, Penna G, Minetti LJ, Presky DH, et al. The role of Stat4 in species-specific regulation of Th cell development by type I IFNs. J Immunol 1998; 161:6567 - 74; PMID: 9862683
- Miyagi T, Takehara T, Nishio K, Shimizu S, Kohga K, Li W, et al. Altered interferon-alpha-signaling in natural killer cells from patients with chronic hepatitis C virus infection. J Hepatol 2010; 53:424 - 30; http://dx.doi.org/10.1016/j.jhep.2010.03.018; PMID: 20554341
- Edlich B, Ahlenstiel G, Zabaleta Azpiroz A, Stoltzfus J, Noureddin M, Serti E, et al. Early changes in interferon signaling define natural killer cell response and refractoriness to interferon-based therapy of hepatitis C patients. Hepatology 2012; 55:39 - 48; http://dx.doi.org/10.1002/hep.24628; PMID: 21898483
- Gough DJ, Messina NL, Clarke CJ, Johnstone RW, Levy DE. Constitutive type I interferon modulates homeostatic balance through tonic signaling. Immunity 2012; 36:166 - 74; http://dx.doi.org/10.1016/j.immuni.2012.01.011; PMID: 22365663
- Kolumam GA, Thomas S, Thompson LJ, Sprent J, Murali-Krishna K. Type I interferons act directly on CD8 T cells to allow clonal expansion and memory formation in response to viral infection. J Exp Med 2005; 202:637 - 50; http://dx.doi.org/10.1084/jem.20050821; PMID: 16129706
- Cui W, Liu Y, Weinstein JS, Craft J, Kaech SM. An interleukin-21-interleukin-10-STAT3 pathway is critical for functional maturation of memory CD8+ T cells. Immunity 2011; 35:792 - 805; http://dx.doi.org/10.1016/j.immuni.2011.09.017; PMID: 22118527
- Chapgier A, Boisson-Dupuis S, Jouanguy E, Vogt G, Feinberg J, Prochnicka-Chalufour A, et al. Novel STAT1 alleles in otherwise healthy patients with mycobacterial disease. PLoS Genet 2006; 2:e131; http://dx.doi.org/10.1371/journal.pgen.0020131; PMID: 16934001
- Chapgier A, Wynn RF, Jouanguy E, Filipe-Santos O, Zhang S, Feinberg J, et al. Human complete Stat-1 deficiency is associated with defective type I and II IFN responses in vitro but immunity to some low virulence viruses in vivo. J Immunol 2006; 176:5078 - 83; PMID: 16585605
- Dupuis S, Dargemont C, Fieschi C, Thomassin N, Rosenzweig S, Harris J, et al. Impairment of mycobacterial but not viral immunity by a germline human STAT1 mutation. Science 2001; 293:300 - 3; http://dx.doi.org/10.1126/science.1061154; PMID: 11452125
- Dupuis S, Jouanguy E, Al-Hajjar S, Fieschi C, Al-Mohsen IZ, Al-Jumaah S, et al. Impaired response to interferon-alpha/beta and lethal viral disease in human STAT1 deficiency. Nat Genet 2003; 33:388 - 91; http://dx.doi.org/10.1038/ng1097; PMID: 12590259
- Liu L, Okada S, Kong XF, Kreins AY, Cypowyj S, Abhyankar A, et al. Gain-of-function human STAT1 mutations impair IL-17 immunity and underlie chronic mucocutaneous candidiasis. J Exp Med 2011; 208:1635 - 48; http://dx.doi.org/10.1084/jem.20110958; PMID: 21727188
- Holland SM, DeLeo FR, Elloumi HZ, Hsu AP, Uzel G, Brodsky N, et al. STAT3 mutations in the hyper-IgE syndrome. N Engl J Med 2007; 357:1608 - 19; http://dx.doi.org/10.1056/NEJMoa073687; PMID: 17881745
- Minegishi Y, Saito M, Tsuchiya S, Tsuge I, Takada H, Hara T, et al. Dominant-negative mutations in the DNA-binding domain of STAT3 cause hyper-IgE syndrome. Nature 2007; 448:1058 - 62; http://dx.doi.org/10.1038/nature06096; PMID: 17676033
- de Beaucoudrey L, Puel A, Filipe-Santos O, Cobat A, Ghandil P, Chrabieh M, et al. Mutations in STAT3 and IL12RB1 impair the development of human IL-17-producing T cells. J Exp Med 2008; 205:1543 - 50; http://dx.doi.org/10.1084/jem.20080321; PMID: 18591412
- Casanova JL, Holland SM, Notarangelo LD. Inborn errors of human JAKs and STATs. Immunity 2012; 36:515 - 28; http://dx.doi.org/10.1016/j.immuni.2012.03.016; PMID: 22520845
- Siegel AM, Heimall J, Freeman AF, Hsu AP, Brittain E, Brenchley JM, et al. A critical role for STAT3 transcription factor signaling in the development and maintenance of human T cell memory. Immunity 2011; 35:806 - 18; http://dx.doi.org/10.1016/j.immuni.2011.09.016; PMID: 22118528