Abstract
Several bispecific antibody-based formats have been developed over the past 25 years in an effort to produce a new generation of immunotherapeutics that target two or more disease mechanisms simultaneously. One such format, the dual-variable domain immunoglobulin (DVD-Ig™), combines the target binding domains of two monoclonal antibodies via flexible naturally occurring linkers, which yields a tetravalent IgG - like molecule. We report the structure of an interleukin (IL)12-IL18 DVD-Ig™ Fab (DFab) fragment with IL18 bound to the inner variable domain (VD) that reveals the remarkable flexibility of the DVD-Ig™ molecule and how the DVD-Ig™ format can function to bind four antigens simultaneously. An understanding of how the inner variable domain retains function is of critical importance for designing DVD-Ig™ molecules, and for better understanding of the flexibility of immunoglobulin variable domains and linkers, which may aid in the design of improved bi- and multi-specific biologics in general.
Keywords: :
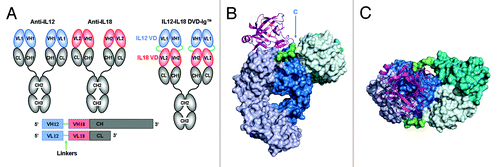
The bispecific tetravalent immunoglobulin known as the dual variable domain immunoglobulin or DVD-Ig™ molecule was first described by Wu, et al. in 2007.Citation1 Like a conventional IgG, the DVD-Ig™ molecule is composed of two heavy chains and two light chains. Unlike IgG, however, both heavy and light chains of a DVD-Ig™ molecule contain an additional variable domain (VD) connected via a linker sequence at the N-termini of the VH and VL of an existing monoclonal antibody (mAb). Thus, when the heavy and the light chains combine, the resulting DVD-Ig™ molecule contains four antigen recognition sites (). The outermost or N-terminal variable domain is termed VD1 and the innermost variable domain is termed VD2; the VD2 is proximal to the C-terminal CH1 or CL. We and others have previously reported that DVD-Ig™ molecules can be manufactured and purified to homogeneity in large quantities, have pharmacological properties similar to those of a conventional IgG1, and show in vivo efficacy in multiple mouse models.Citation1,Citation2
Figure 1. DVD-Ig™ Technology Overview. (A) A DVD-Ig™ binding protein is constructed from two parent antibodies by addition of the first variable domain to the second via a flexible linker sequence (in this case antiIL12 antiIL18 and SS linkers). (B) Solid surface representation of IL12-IL18 DVD-Ig™ DFab with IL18 shown as ribbon. VD1 is shown in teal and VD2 and constant domains are shown in blue. Heavy and light chains are dark and light shades respectively. Short-short linkers are shown in green. IL18 is shown in magenta. (C) View of b from the top.
Theoretically, the structurally novel DVD-Ig™ format design can impose certain structural and functional constraints on VD2, the inner variable domain. In a conventional mAb, the complementarity-determining regions (CDRs) of a VD are surface exposed with no N-terminal constraints, no limitations as to the size and location (soluble or cell surface) of the target antigen, and no constraints as to conformational changes that might occur upon target binding (VD stabilization). In the DVD-Ig™ molecule, the juxtaposition of VD1 to VD2 via linkers could potentially occlude VD2 CDRs, limit VD2 rotational flexibility, impose limits on target size and location, or impose constraints on VD2 conformational changes (stabilization) upon target binding. We and others, however, have observed that affinity at the inner antigen binding site (VD2) may be somewhat dependant on the VD1/VD2 pair combination (amino acid sequences), the VD1/VD2 orientation, and linker selection.Citation2-Citation4 The choice of linker length between the VD1 and VD2 (e.g., either both short linkers [S-S], both long linkers [L-L], or one short and one long linker [S-L or L-S] as shown in ), can affect the affinity of the inner VD2 domain.Citation4 In addition, the antigen affinity at the outer antigen binding domain (VD1) is often nearly equal to the parent antibody in the DVD-Ig™ format. Both VD1 and VD2 of a DVD-Ig™ molecule can successfully target soluble and cell surface antigens and can bind antigens simultaneously and with full occupancy.Citation1 Here, we report the structure of the interleukin (IL)12-IL18 DVD-Ig™ Fab with IL18 bound at VD2. The structure provides the basis for understanding how the DVD-Ig™ molecule binds two different antigens simultaneously and lays the foundation for hypothesis-driven design of new DVD-Ig™ molecules with adjustable binding properties.
Table 1. DVD-Ig™ linkers
An initial view of the DVD-Ig™ DFab structure immediately reveals how the DVD-Ig™ molecule functions to bind two different antigens on each DFab simultaneously (). With IL18 bound to VD2 (the inner variable domain), the outer VD1 rests entirely on top of the heavy chain of the inner variable domain. This orientation of the outer variable domain positions the CDRs of VD1 for binding the second antigen approximately 85° from the inner antigen CDRs (), leaving ample room for binding the outer antigen (IL12). The structure helps explain how the binding affinity for both antigens in this DVD-Ig™ molecule remains essentially unchanged from the parent antibodies [KD (pM) mAbs: IL12 = 120, IL18 = 140; IL12-IL18 DVD-Ig™: IL12 = 130, IL18 = 160].Citation1 Inspection of the structure from the top of the DVD-Ig™ molecule reveals no apparent contact between IL18 and the outer VD1(), and no significant interactions between the linkers and the inner VD2 CDRs or the outer VD1. Viewing the structure from the side and looking underneath the outer VD1 (), there appears to be a surface complementarity between the inner surface of the outer VD1 and the outer surface of the VD2 heavy chain, and there is an approximate 10° twist to the outer VD with respect to the inner VD. When the outer VD1 rests upon the VD2 heavy chain surface, it results in an overall additional buried surface area of 1745Å2.Citation5 The surface complementarity statistic, Sc = 0.67 for the interface between the outer VD2 and the inner VD1 heavy chain indicates considerable surface complementarity, but also suggests that the interface can be significantly improved.Citation6 Adjusting the linker sequences may help situate the outer variable domain, VD1, in a more favorable position where the surface complementarity statistic and the overall buried surface area would be greater.
Figure 2. Structure of IL12-IL18 DVD-Ig™ DFab fragment with IL18 bound. (A) Ribbon and solid surface representation of the structure with heavy and light chains in respective shades of gray, linkers in green, and IL18 in magenta. The epitope of IL18 (dark blue sticks) interacts mainly with heavy chain CDRs 1 (lt. cyan) and 3 (lt. orange) with minor contributions from heavy and light chain CDR 2 (lt. yellow). (B) The VD2 CDR plane as viewed from above and in ribbon with IL18 removed for viewing clarity (C) Ribbon representation viewed from below the outer VD. Heavy chain is shown in dark blue and light chain in teal. (D) Stereo diagram of a representative portion of the electron density map at the DFab IL18 VD/IL18 interface.
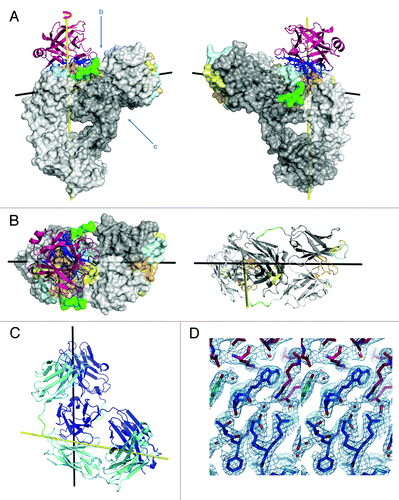
In the context of the DVD-Ig™ format, we are able to identify critical amino acid interactions as well as water mediated hydrogen bonds that are responsible for producing the (140pM) KD for the IL18 parental antibody. It is worth noting that, until now, we have been unsuccessful at producing crystals of this particular IL18 Fab (derived from the parental IgG) complexed with IL18 at a sufficient quality for X-ray crystal structure determination. Some of the critical interactions made by the IL18 Fv to IL18 are shown along with a representative portion of the electron density map in . The surface complementarity statistic is Sc = 0.73 for the interface between IL18 and the IL12-IL18 DFab, which is about average for antibody antigen complexes, and is coincidentally identical to the previously reported value for the IL18/125–2H Fab interfaceCitation6,Citation7 that recognizes an entirely different epitope. Comparing the KD of the parent anti-IL18 antibody (KD = 140pM) to the KD of the IL12-IL18 DVD-Ig™ for IL18 (KD = 160pM), it is clear that there is little loss of affinity for the binding of IL18 at the inner antigen binding site (VD2) in the DVD-Ig™ architecture. Additionally, superposition of the parental IL18 Fab (2VXV)Citation6 onto the IL18 Fab portion of the IL12-IL18 DFab reveals a low main chain rmsd = 0.334 Å.
The ‘short-short’ linkers () were used for the construction of IL12-IL18 DVD-Ig™ and are composed of sequences taken proximal to the elbow regions of a conventional IgG1. They directly connect the C-terminus of the outer IL12 VD1 to the N-terminus of the inner IL18 VD2 of the molecule, which results in a tetravalent, dual specific, molecule with full activity for both antigens compared with the parental IgGs. The light chain linker sequence is TVAAP and the heavy chain linker sequence is ASTKGP.Citation1 We see complete electron density for the light chain linker, but we only see main chain density for the heavy chain linker because it is more solvent exposed. The outer variable domain can be imagined as a swinging bucket that can move over the inner antigen binding site from side to side with the linkers acting as flexible handles connecting it to the mAb Fab portion below. This outer VD “bucket” concept suggests that we may affect the outer VD movement by adjusting linker lengths and thereby modulate inner domain binding.
Having two linkers to the outer variable domain in the DVD-Ig™ format affords additional flexibility when engineering DVD-Ig™ molecules. By adjusting the length and sequence of the linkers, the position of the outer variable domain may be altered, making it possible to tune the affinity of the inner antigen site. A cartoon schematic illustrating how varying the linker length can reposition the outer VD to improve interactions with the inner VD is shown in . We have seen a significant loss of affinity for vascular endothelial growth factor (VEGF) when it is expressed at the inner VD2 with SS linkers (Table S2). To investigate the effect of linker length and outer VD sequence pairing on inner VD antigen affinity, we constructed a matrix of DVD-Ig™ proteins pairing anti-VEGF VD with four different paratopes of an alternate therapeutic target of 54.7 kDa that we refer to as AgA. These DVD-Ig™ molecules were made with four different linker sets, short-short (SS), short-long (SL), long-short (LS), and long-long (LL), as described in Table S2. The VEGF binding kinetics for this series of DVD-Ig™ proteins suggest that the affinity for the inner antigen can be modulated by adjusting linker length, and that it is possible to screen for desired linker length combinations. The binding kinetics () reveal a linker length dependent reduction of VEGF affinity (KD) for the inner VD that is primarily the result of reduced on-rate (ka) for short linkers, relative to the reference IgG. Furthermore, the binding kinetics show that the KD can be modulated by adjusting linker length and by using different outer VD sequences. Overall, we can always identify a DVD-Ig™ molecule retaining binding affinities to both inner and outer variable domains through a process of selecting the best inner/outer variable domain combination, optimizing the orientation of the two variable domains and adjusting the linker length and type. The ability to modulate the affinity of one antigen with respect to the other may be an important feature for any dual specific binding protein when dosing two paratopes simultaneously. Although it is not necessary for the proper function of a DVD-Ig™ molecule, in certain applications one or both of the linkers can be particularly engineered to be enzymatically cleaved to enhance or activate antigen binding at the inner antigen site. Cleavage of one linker allows outer VD1 additional rotational freedom, and greater accessibility to the inner VD2 antigen binding site. This feature may enable tissue targeting of the DVD-Ig™ molecule with the outer variable domain while masking the paratope of the inner domain until the DVD-Ig™ molecule is “activated” at the targeted site by tissue specific proteases. We have recently shown that a DVD-Ig™ molecule that substantially lost antigen affinity at the inner VD2 binding site can regain binding affinity to near that of the parent antibody while still maintaining the binding functionality of the outer VD1 after selective cleavage of one of the linkers.Citation3
Figure 3. Linker Length Affects Inner Antigen Affinity. (A) Illustration of outer VD position as a function of linker length, relative to the inner VD. Arrows indicate likely reorientation of the outer VD. (B) The VEGF and AgA binding kinetics are shown in plots of off-rate (kd) vs. on-rate (ka). Symbols are defined in ; briefly, data points are colored by linker set (red, green, dark blue and brown correspond to linker sets SS, LS, SL and LL, respectively) and shaped by antiAgA sequence (circle, triangle, square and diamond are sequences 1, 2, 3 and 4, respectively). Binding curves with fits, for both VEGF and AgA, as well as the detailed methodology, can be found in supplemental materials.
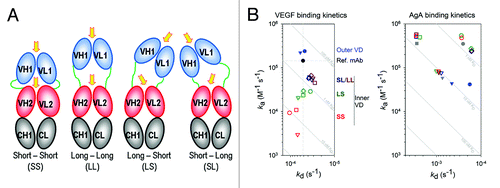
Table 2. DVD-Ig™ binding kinetics for AgA and VEGF
The recent approval of the first bispecific therapeutic mAb (catumaxomab) in the EU,Citation8 emerging clinical data,Citation9 and entry of several bispecifics into clinical trials has renewed interest in design and development of bi- and multi-specific biologics. In addition, bispecific antibodies offer novel opportunities not possible with mAbs, such as recruitment of specific immune cell populations to kill tumors,Citation8,Citation9 targeting biologics across the blood brain barrier,Citation10 and delivering therapeutic agents, including stem cells, to specific sites.Citation11,Citation12 Challenges in developing therapeutic bispecifics, however, remain, e.g., molecular stability, pharmacokinetics, manufacturability of individual bispecific formats. Of the more than 30 bispecific formats described to date, there are no crystal structures reported for any other bispecific format that reveals both paratopes with an antigen bound. This fact speaks to the robust nature of the DVD-Ig™ format and the ability to produce these proteins in large quantities in a highly purified and stable form. The DVD-Ig™ format is a novel immunotherapeutic platform that exploits the benefits of the immunoglobulin fold through optimization of its various components while engineering in the necessary flexibility to bind an additional antigen. This platform can be utilized to bind a combination of both soluble or cell surface expressed antigens simultaneously. The information contained within the structure of the complex of the DFab with IL18 has proven to be particularly useful for understanding antigen binding properties of other DVD-Ig™ protein pairs and for designing future DVD-Ig™ molecules. There is obviously a need for additional structural studies with a variety of antigens at both the inner and outer variable domain positions in order to more fully comprehend the dynamic nature of the DVD-Ig™, in particular how the various states of antigen occupancy affect VD positioning and how the different antibody frameworks interact with one another and reposition upon antigen binding. It is expected that this structure and future structures will facilitate the design of a new generation of dual specific therapeutics.
Materials and Methods
Protein production, purification and crystallization
All DVD-Ig™ molecules were prepared as previously described.Citation4 DFab was prepared from the DVD-IgTM 4 molecules by standard papain digestion and purified using high performance ion exchange and gel filtration. A 3-fold excess of IL18 was used to make the complex. The complex was separated from excess IL18 by gel filtration, concentrated to 17mg/ml, and crystallized by vapor diffusion in a 1:1 ratio against a reservoir containing 1mL of 2M ammonium sulfate, 0.1M sodium acetate pH 4.6 at 4°C. Crystals were flash frozen in liquid nitrogen using reservoir solution and 25% (v/v) glycerol.
X-ray data collection, structure solution and analysis
Data were collected at 100K using 1Å wavelength at the Advanced Light Source 502 beamline to 2.8Å and processed using HKL2000.Citation13 Initial phases were obtained by molecular replacement using the Fab, and VD from pdb code 2VXVCitation7 and IL-18 from pdb code 2VXTCitation7 using the programs PhaserCitation14 and MolRepCitation15 (CCP4Citation16 suite of programs). A randomly selected set of 5% of total reflections was used for Rfree calculations. Iterative cycles of refinement and model building were performed using Refmac,Citation17 AutoBuster,Citation18 and Coot.Citation19,Citation20 A second data set was collected in a similar fashion using the Advanced Photon Source IMCA 17-ID beamline on a single crystal which grew after 22 mo and diffracted to 2.1Å. The data were processed using AutoProcCitation21 and refined as above. The quality of the final structure was evaluated using programs within the CCP4 suite with 98.7% of all residues in favored or allowed regions of the Ramachandran diagram. Final refinement statistics can be found in Table S1. All alignments were performed using SSM superpositioning in Coot;Citation19,Citation20 electron density maps were calculated using CCP4,Citation16 the surface areas and complementarity statistics were calculated with the programs AREAIMOL and SCCitation5 respectively (CCP4Citation16 program suite); and all molecular figures were generated using the program PyMol.Citation22
Binding analysis
Protocols for SPR binding studies and individual sensograms are available in the online supplementary information.
Accession code
The X-ray crystallographic coordinates have been deposited in the Protein Data Bank with accession ID 4HJJ.
| Abbreviations: | ||
| DVD-Ig™ | = | Dual-Variable Domain immunoglobulin™ molecule |
| mAb | = | monoclonal antibody |
| IgG | = | Immunoglobulin G |
| Fab | = | Antigen binding Fragment of an immunoglobulin |
| DFab | = | DVD-Ig™ Fab fragment |
| IL12 and IL18 | = | Interleukins 12 and 18 |
| VD | = | VD1, and VD2, Variable domain, outer and inner variable domain |
| CH and CL | = | Constant heavy and constant light chain domains |
| VH and VL | = | Variable heavy and constant light chain domains |
| CDRs | = | Complementarity determining regions |
| Sc | = | Surface complementarity statistic |
| rmsd | = | Root-mean-square deviation |
Additional material
Download Zip (940.2 KB)Acknowledgments
We thank Virgina Rath (Reciprocal Space Consulting, L. L. C., Oakland, CA) for the X-ray diffraction data collection for the first data set at the Advanced Light Source, Berkeley, CA. Final X-ray diffraction data were collected at the facilities of the Industrial Macromolecular Crystallography Association Collaborative Access Team (IMCA-CAT) at the Advanced Photon Source. These facilities are supported by the companies of the Industrial Macromolecular Crystallography Association. We thank Chengbin Wu and Hua Ying formerly of Abbott Bioresearch Center for making the IL12-IL18 DVD-Ig™ construct; Alexander Ivanov, George Cunha, Carrie Goodreau, David Lee, and Georgeen Gaza-Bulseco of AbbVie Bioresearch Center for their contributions to preliminary DVD-Ig™ analyses; Diana Steel of AbbVie Bioresearch Center for her counsel and assistance in preparing the manuscript; and for support for the DVD-Ig™ initiative, manuscript review, and feedback we thank Peter Isakson, and Jochen Salfeld of AbbVie Bioresearch Center, Jonathan Greer, Karl Walter, and Vincent Stoll of AbbVie.
Disclosure of Potential Conflicts of Interest
The work presented in this manuscript was fully funded by AbbVie, Inc. and the authors are employees of AbbVie, Inc. (formerly Abbott Laboratories), and may own company stock or stock options.
Author Contributions
R.E. purified the protein and prepared the complex. R.J. crystallized the complex. C.J. solved and analyzed the structure. J.G., and Y.L. completed sequence retrieval and design of VEGF-AgA DVD-Ig™’s, and E.D. performed the Biacore studies. C.J., T.G., R.J., and E.D. prepared the manuscript. T.G. leads the DVD-Ig™ Initiative team.
References
- Wu C, Ying H, Grinnell C, Bryant S, Miller R, Clabbers A, et al. Simultaneous targeting of multiple disease mediators by a dual-variable-domain immunoglobulin. Nat Biotechnol 2007; 25:1290 - 7; http://dx.doi.org/10.1038/nbt1345; PMID: 17934452
- Kou G, Shi J, Chen L, Zhang D, Hou S, Zhao L, et al. A bispecific antibody effectively inhibits tumor growth and metastasis by simultaneous blocking vascular endothelial growth factor A and osteopontin. Cancer Lett 2010; 299:130 - 6; http://dx.doi.org/10.1016/j.canlet.2010.08.011; PMID: 20826049
- Digiammarino EL, Harlan JE, Walter KA, Ladror US, Edalji RP, Hutchins CW, et al. Ligand association rates to the inner-variable-domain of a dual-variable-domain immunoglobulin are significantly impacted by linker design. MAbs 2011; 3:487 - 94; http://dx.doi.org/10.4161/mabs.3.5.16326; PMID: 21814039
- Wu C, Ying H, Bose S, Miller R, Medina L, Santora L, et al. Molecular construction and optimization of anti-human IL-1alpha/beta dual variable domain immunoglobulin (DVD-Ig) molecules. MAbs 2009; 1:339 - 47; http://dx.doi.org/10.4161/mabs.1.4.8755; PMID: 20068402
- Winn MD. An overview of the CCP4 project in protein crystallography: an example of a collaborative project. J Synchrotron Radiat 2003; 10:23 - 5; http://dx.doi.org/10.1107/S0909049502017235; PMID: 12511787
- Lawrence MC, Colman PM. Shape complementarity at protein/protein interfaces. J Mol Biol 1993; 234:946 - 50; http://dx.doi.org/10.1006/jmbi.1993.1648; PMID: 8263940
- Argiriadi MA, Xiang T, Wu C, Ghayur T, Borhani DW. Unusual water-mediated antigenic recognition of the proinflammatory cytokine interleukin-18. J Biol Chem 2009; 284:24478 - 89; http://dx.doi.org/10.1074/jbc.M109.023887; PMID: 19553661
- Heiss MM, Murawa P, Koralewski P, Kutarska E, Kolesnik OO, Ivanchenko VV, et al. The trifunctional antibody catumaxomab for the treatment of malignant ascites due to epithelial cancer: Results of a prospective randomized phase II/III trial. Int J Cancer 2010; 127:2209 - 21; http://dx.doi.org/10.1002/ijc.25423; PMID: 20473913
- Topp MS, Kufer P, Gökbuget N, Goebeler M, Klinger M, Neumann S, et al. Targeted therapy with the T-cell-engaging antibody blinatumomab of chemotherapy-refractory minimal residual disease in B-lineage acute lymphoblastic leukemia patients results in high response rate and prolonged leukemia-free survival. J Clin Oncol 2011; 29:2493 - 8; http://dx.doi.org/10.1200/JCO.2010.32.7270; PMID: 21576633
- Yu YJ, Zhang Y, Kenrick M, Hoyte K, Luk W, Lu Y, et al. Boosting brain uptake of a therapeutic antibody by reducing its affinity for a transcytosis target. Sci Transl Med 2011; 3:84ra44; http://dx.doi.org/10.1126/scitranslmed.3002230; PMID: 21613623
- Metz S, Haas AK, Daub K, Croasdale R, Stracke J, Lau W, et al. Bispecific digoxigenin-binding antibodies for targeted payload delivery. Proc Natl Acad Sci U S A 2011; 108:8194 - 9; http://dx.doi.org/10.1073/pnas.1018565108; PMID: 21536919
- Zhao TC, Tseng A, Yano N, Tseng Y, Davol PA, Lee RJ, et al. Targeting human CD34+ hematopoietic stem cells with anti-CD45 x anti-myosin light-chain bispecific antibody preserves cardiac function in myocardial infarction. J Appl Physiol 2008; 104:1793 - 800; http://dx.doi.org/10.1152/japplphysiol.01109.2007; PMID: 18292296
- Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol 1997; 276:307 - 26; http://dx.doi.org/10.1016/S0076-6879(97)76066-X
- McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J Appl Crystallogr 2007; 40:658 - 74; http://dx.doi.org/10.1107/S0021889807021206; PMID: 19461840
- Vagin A, Teplyakov A. An approach to multi-copy search in molecular replacement. Acta Crystallogr D Biol Crystallogr 2000; 56:1622 - 4; http://dx.doi.org/10.1107/S0907444900013780; PMID: 11092928
- Collaborative Computational Project, Number 4. The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr 1994; 50:760 - 3; http://dx.doi.org/10.1107/S0907444994003112; PMID: 15299374
- Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr 1997; 53:240 - 55; http://dx.doi.org/10.1107/S0907444996012255; PMID: 15299926
- Blanc E, Roversi P, Vonrhein C, Flensburg C, Lea SM, Bricogne G. Refinement of severely incomplete structures with maximum likelihood in BUSTER-TNT. Acta Crystallogr D Biol Crystallogr 2004; 60:2210 - 21; http://dx.doi.org/10.1107/S0907444904016427; PMID: 15572774
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 2004; 60:2126 - 32; http://dx.doi.org/10.1107/S0907444904019158; PMID: 15572765
- Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr 2010; 66:486 - 501; http://dx.doi.org/10.1107/S0907444910007493; PMID: 20383002
- Vonrhein C, Flensburg C, Keller P, Sharff A, Smart O, Paciorek W, et al. Data processing and analysis with the autoPROC toolbox. Acta Crystallogr D Biol Crystallogr 2011; 67:293 - 302; http://dx.doi.org/10.1107/S0907444911007773; PMID: 21460447
- Schrodinger LLC. . (2010).