Abstract
This report describes the design, generation and testing of Ylanthia, a fully synthetic human Fab antibody library with 1.3E+11 clones. Ylanthia comprises 36 fixed immunoglobulin (Ig) variable heavy (VH)/variable light (VL) chain pairs, which cover a broad range of canonical complementarity-determining region (CDR) structures. The variable Ig heavy and Ig light (VH/VL) chain pairs were selected for biophysical characteristics favorable to manufacturing and development. The selection process included multiple parameters, e.g., assessment of protein expression yield, thermal stability and aggregation propensity in fragment antigen binding (Fab) and IgG1 formats, and relative Fab display rate on phage. The framework regions are fixed and the diversified CDRs were designed based on a systematic analysis of a large set of rearranged human antibody sequences. Care was taken to minimize the occurrence of potential posttranslational modification sites within the CDRs. Phage selection was performed against various antigens and unique antibodies with excellent biophysical properties were isolated. Our results confirm that quality can be built into an antibody library by prudent selection of unmodified, fully human VH/VL pairs as scaffolds.
Introduction
Due to their high specificity and broad target space, monoclonal antibodies (mAbs) represent the most important class of biologics and have great potential for both diagnostic and therapeutic applications.Citation1 A diverse set of in vivo and in vitro technologies are currently available for the generation of these versatile molecules. Fully human mAbs, for example, can be derived from human B cells by immortalization via viral transformation,Citation2 through the generation of human hybridomas,Citation3 or through direct cloning of Ig encoding gene transcripts.Citation4-Citation6 Target specific human B cell material, however, in some cases may not be readily accessible. The hybridoma technologyCitation7 using immunoglobulin (Ig) transgenic mouse systemsCitation8 overcomes this limitation but still suffers from the restriction of the target space due to the need for immunogenic sequences and epitopes. On the other hand, the mentioned in vivo methods share the advantage of delivering structurally diverse IgM, IgG and IgA antibodies with naturally paired and correctly folded variable Ig heavy and Ig light (VH/VL) chain frameworks.
Among the in vitro technologies, the methods of choice for the generation of human mAbs are phage, ribosome and yeast display of recombinant antibody libraries.Citation9-Citation14 Various naïve,Citation15,Citation16 immune,Citation17 semi-syntheticCitation18 and syntheticCitation19-Citation21 library formats have been described. They differ both in their design regarding framework composition and complementarity-determining region (CDR) diversification,Citation22 as well as their quality, as defined by library size and correct sequences. Major advantages of phage display include the virtual independence of the selection process from target characteristics (e.g., immunogenicity or toxicity of the antigen), which allows application to a wide range of antigens, the possibility of accessing rodent cross reactive antibodies for animal models, the full control over the selection conditions and the technological robustness and relative speed.Citation23
Therapeutic antibodies must fulfill high standards with regard to their binding affinity, target and epitope specificity, and functional activity. An additional challenge of antibody discovery technologies is the generation of mAbs with appropriate biophysical properties. To reach the market, antibody therapeutics ideally should, for example, not aggregate or precipitate and resist degradation by proteases, as these factors contribute to optimal manufacturing and shelf life properties and ensure a long serum half-life. Furthermore, the increasing demands of regulatory authorities with respect to properties such as the homogeneity of the product drive these needs. Similar stability requirements also apply for diagnostic and research mAb tools.Citation24
Various approaches have been undertaken to combine the advantages of in vivo and in vitro technologies to generate recombinant libraries that include predominantly antibodies with superior properties. For example, the human combinatorial antibody library (HuCAL) platform is based on a composition of selected VH and VL consensus master gene sequences that are diversified using modular CDR cassettes.Citation19 HuCAL PLATINUM, the latest generation HuCAL antibody library,Citation20 implemented optimized antibody framework sequences with a reduction of unproductive sequences and a revised rational CDR design compared with the previous HuCAL scFv and HuCAL GOLD libraries.Citation19,Citation21 Another approach to combine structural CDR diversity with a biophysically optimal VH/VL combination is the n-CoDeR scFv antibody library, which uses a VH3–23/Vλ1–47 pairing as scaffold.Citation25
The Ylanthia antibody library introduces a new concept in which fixed VH/VL framework pairs are rationally selected based on biophysical characteristics that are relevant for antibody selection, development and manufacturing. Through this process, optimal biophysical properties can be built into the library using human germline antibody sequences, without the need to introduce stabilizing mutations. The work-flow leading to the final framework composition of the library comprised several steps. First, 20 VH, 12 Vκ and 8 Vλ Ig gene segments were chosen based on their prevalence in the human antibody repertoire and additional in silico analysis to cover all canonical CDR structures. The 400 VH/VL pairings resulting from the combination of those 40 variable domains were characterized in vitro to rank VH/VL framework pairs according to properties, such as relative protein expression level, thermal and serum stability, aggregation propensity in Fab and IgG1 format and relative Fab CysDisplay levels on phage. At the end of this testing, 36 fixed and distinct VH/VL framework pairs were selected as variable region scaffolds for Ylanthia. Within these 36 VH/VL framework pairs, high structural diversity is maintained as important basis for selecting antibodies against a broad range of different epitopes and target classes.
The CDR-H1 and CDR-H2 design of these 36 VH/VL framework pairs was based on germline-encoded CDR sequences. Diversity was introduced to remove potential post-translational modification sites (PTMs) (e.g., deamidation sites Asn-Ser, NS; Asn-Gly, NG; Asn-His NH; isomerization sites Asp-Ser, DS; Asp-Gly, DG; Asp-Asp,DD; protease cleavage sites Asp-Pro, DP; Asp-Glu, DQ; oxidation sites Met, M)Citation26 within CDR-H1 and CDR-H2 in a significant proportion of the library. To enhance antibody Fab expression in bacterial and IgG expression in mammalian systems, nucleotide sequence codon optimization was implemented during library construction.Citation27 The use of the TRIMCitation28 and the SlonomicsCitation29 technology to engineer diversified CDR-L3 and CDR-H3 cassettes, respectively, allowed for sophisticated CDR design and guarantees a very high proportion of correct clones. The amino acid diversity of the CDR3 cassettes was based on the Ig gene segment- (CDR-L3) and on the CDR length (CDR-H3) to mimic the natural position-dependent occurrence of amino acids in the CDR3s with a strong reduction or omission of potential PTMs.
To confirm the functionality and quality of the Ylanthia antibody library, phage selections on recombinant human (rh) TNF-α, M-CSF, rhErbB4, rhFZD-4, eGFP and several other antigens were performed. Selected Fab fragments were converted into full-length human IgG1 antibodies that were characterized for their biophysical properties. Our results show that the 36 VH/VL pairs that compose Ylanthia translate into highly diverse sets of target-specific and high affinity antibodies with favorable biophysical characteristics.
Results
Analysis of the human antibody repertoire
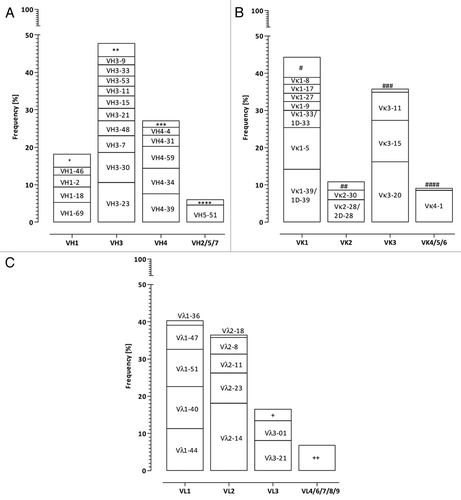
To assess the relative frequency of VH and VL frameworks and to identify preferentially occurring pairings in natural antibody repertoires, published and in-house generated data were evaluated in detail. Single B cell studies utilizing samples from both healthy and autoimmune subjectsCitation30-Citation35 were analyzed, and data from naïve unmutated IgM (n = 1348), somatically mutated IgM (n = 158) and IgG (n = 632) antibodies were combined to determine the most prevalent VH/VL pairings. In addition, Ig gene transcript information was gained experimentally from single mature naïve B cells and antibody secreting cells isolated from a healthy donor. In total, 2138 VH, 1358 Vκ and 780 Vλ functionally rearranged sequences were included in the analysis of VH/VL pairing frequency. Additional, 326 VHCitation36,Citation37 and 298 VκCitation38 productive sequences were utilized in the ranking of the most frequently used VH and Vκ gene segments.
VH3–23 (10.6%) and VH3–30 (8.0%) gene segments were found to be the most frequent among the analyzed human VH sequences, followed by three members of the VH4 gene family, VH4–39 (7.6%), VH4–34 (6.8%) and VH4–59 (5.8%) as well as VH1–69 (5.3%). Within the human Vκ sequences Vκ3–20, Vκ1–39, Vκ1–5, Vκ3–15 and Vκ4–1 together constituted more than 60% of all analyzed Vκ sequences. Vλ2–14 was the most frequently found Vλ gene segment (18.1%), followed by Vλ1–40 (11.3%), Vλ1–44 (11.3%) and Vλ1–51 (10.0%) ().
Figure 1. Frequency distribution of (A) VH, (B) Vκ and (C) Vλ gene segments in the natural human antibody repertoire. V gene usage for human antibodies was compiled from several B cell repertoire studies Citation30-Citation38 and own sequencing efforts as described in the methods section. The names of the most frequent V segments are listed in the respective boxes, less frequent used V segment groups are indicated by *(VH1–3, VH1–8, VH1–24, VH1–58), **(VH3–13, VH3–20, VH3–43, VH3–49, VH3–64, VH3–66, VH3–72, VH3–73, VH3–74), ***(VH4–28, VH4–30.2, VH4–61), ****(VH2–5, VH6–1, VH7–4.1, VH7–81), #(Vκ1–6, Vκ1D-8, Vκ1–12/1D-12, Vκ1–13/1D13, Vκ1–16/1D16, Vκ1D-17, Vκ1D-43), ##(Vκ2–24, Vκ2–29, Vκ2–40/2D-40, Vκ2D-29, Vκ2D-30), ###(Vκ3D-7, Vκ3D-11, Vκ3D-15, Vκ3D-20), ####(Vκ5–2, Vκ6–21/6D-21, Vκ6D-41), +(Vλ3–9, Vλ3–10, Vλ3–12, Vλ3–19, Vλ3–22, Vλ3–25, Vλ3–27) and ++(Vλ4–60, Vλ4–69, Vλ6–57, Vλ7–43, Vλ7–46, Vλ8–61, Vλ9–49).
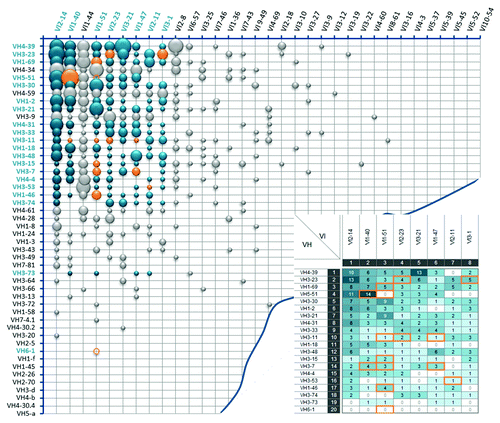
The most prevalent natural VH/VL pairs were primarily composed of the most frequently occurring VH and VL gene segments. This observation correlates with previous reports, which showed no obvious indication for a preferential pairing of certain germline sequences.Citation39 Within the combined Ig kappa () and Ig lambda () antibody repertoire analyzed, a total of 694 different VH/VL pairs (383 VH/Vκ and 311 VH/Vλ) out of approximately 3,400 theoretically possible VH/VL combinations have been found.
Figure 2. Frequency distribution of VH/Vκ antibody pairs in the natural human antibody repertoire. Bubble chart illustrating VH/Vκ pairing frequencies of 1,358 analyzed VH/Vκ sequences from the natural human antibody repertoire. V segments are listed according to their individual frequency in the Ig kappa antibody pool. Each bubble size refers to the relative frequency of the corresponding VH/Vκ pair. Green bubbles indicate the 240 selected VH/Vκ pairings for the initial screening experiments, with their absolute frequencies summarized in the inlay picture. Orange bubbles, circles and frames show the final selected 21 VH/Vκ pairs that are included in Ylanthia. The circles indicate VH/Vκ pairs that were not found in our analysis of natural VH/VL pair sequences.
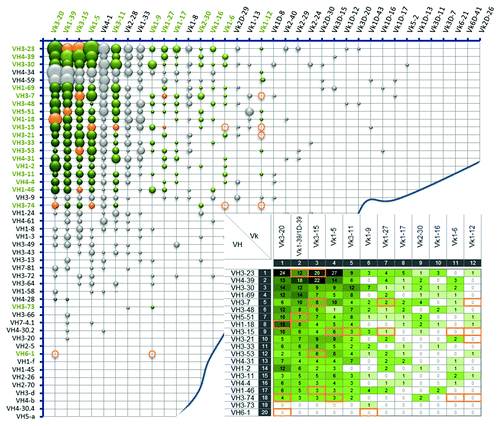
Figure 3. Frequency distribution of VH/Vλ antibody pairs in the natural human antibody repertoire. Bubble chart illustrating VH/Vκ pairing frequencies of 780 analyzed VH/Vλ sequences from the natural human antibody repertoire. V segments are listed according to their individual frequency in the Ig lambda antibody pool. Each bubble size refers to the relative frequency of the corresponding VH/Vλ pair. Blue bubbles indicate the 160 selected VH/Vλ pairings for the initial screening experiments, with their absolute frequencies summarized in the inlay picture. Orange bubbles, circles and frames show the final selected 15 VH/Vλ pairs that are included in Ylanthia. The circles indicate VH/Vλ pairs that were not found in our analysis of natural VH/VL pair sequences.
An in-depth in silico and experimental in vitro analysis was performed to identify germline VH/VL pairs that are ideally suited as frameworks for this novel type of antibody library. While the focus was to identify VH/VL pairs with superior biophysical properties, care was also taken to maintain diversity in Ig V gene segment families and canonical CDR structures.
In silico analysis of frequent germline VH, Vκ and Vλ gene segments
Unpaired VH, Vκ and Vλ protein sequences, which accounted for at least 1.5%, 1% and 5% of the investigated Ig sequence repertoire, respectively, were reviewed in silico in their germline configuration for the following properties: 1) isoelectric point (pI), 2) CDR1 and CDR2 canonical structuresCitation40 and 3) number of potential PTMs and free cysteine (Cys, C) residues (). Not all, however, of the most prominent VH, Vκ and Vλ sequences were selected for this in silico analysis. Concerning the VH repertoire, for example, VH4–34, and VH4–59 were excluded, because VH4-containing antibodies have been found to be heavily de-selected during phage display.Citation19,Citation21,Citation41 Furthermore, antibodies derived from VH4–34 might potentially be cytotoxic to B cells.Citation42 The VH3–9 sequence was omitted because its canonical CDR-H structures were already overrepresented by the other selected VH3 family gene members. In addition, the less frequently occurring VH sequences of VH3–73 and VH6–1 were included to enhance the diversity of canonical CDR-H structures. The prominent Vκ and Vλ gene segments, Vκ4–1, Vκ2–28/2D-28, Vκ1–33/1D-33, Vκ1–8, Vκ2D–29 and Vλ1–44 were also excluded for the reasons described aboveCitation20,Citation43 and to prevent an overrepresentation of Vκ1 and Vλ1 family sequences.
Table 1. In silico analysis of selected V gene element amino acid sequences
The isoelectric point (pI) of a therapeutic antibody is an important aspect for formulation development. Values of pIs in the range of the pHs commonly used for antibody formulation, i.e., in the physiological or slightly acidic range, can be problematic due to a potentially increased propensity of such antibodies to aggregate under these conditions. Within the selected VH gene segment sequences, the calculated pI values ranged from 8.0 (VH3–48) to 9.5 (VH3–73). In the analyzed Vκ gene segment sequence pool the observed pI values were between 5.0 (Vκ1–5) and 8.7 (Vκ1–17, Vκ1–27). The selected Vλ gene segments displayed the lowest pI values ranging from 4.5 (Vλ3–1) to 6.7 (Vλ2–11, Vλ2–23).
All classically defined canonical CDR-H1 (H1–1, H1–2 and H1–3) and CDR-H2 (H2–1, H2–2, H2–3, H2–4 and H2–5) structures,Citation40 as well as various length variants of Vκ and Vλ CDR-L1,Citation44,Citation45 were present in the selection of germline V gene segments (). The majority of the analyzed CDR-H1 and CDR-H2 sequences included potential PTM sites. Of the analyzed germline VH gene segments, only VH4 family members and VH1–69 had no such sites in their CDR-H1 and CDR-H2 sequences. The VH3 family genes, however, exhibited major PTM hot spots. Interestingly, critical Asn-Xaa-Ser, NxS and Asn-Xaa-Thr, NxT sequence motifs for N-linked glycosylation were not present in the germline CDR1 and CDR2 sequences of any of the analyzed VH and VL gene segments. They can, however, be generated during somatic hypermutation.Citation46
Through the in silico analysis described above, a set of 20 VH and 20 VL germline sequences (20 VH, 12 Vκ and 8 Vλ) was selected. The inclusion criteria were frequency of natural occurrence, diversity in Ig V gene segment families, canonical CDR structures, and rational selection based on experience gathered through in-house in vitro antibody generation. In addition, favorable pI values and absence of potential PTM sites were considered to be important aspects as they might hamper later manufacturing and development steps.
In vitro screening of VH/VL pairs in Fab and human IgG1 format
The selected 20 VH, 12 Vκ and 8 Vλ gene segments were combined to generate VH/VL pairs for experimental analysis. A set of predictive screening assays was performed as described below to reduce the number of VH/VL pairs for subsequent, more detailed analysis.
The 400 VH/VL pairs resulting from the combination of the 20 VHs with 20 VLs represent the most prominent VH/VL pairs in the human antibody repertoire ( and ). During testing, all VH regions carried the same CDR-H3, which is derived from the VH region of the hu4D5–8 antibody,Citation19,Citation47 and all VL regions carry a kappa-like and lambda-like CDR-L3, respectively.Citation48
Relative expression yields of Fab and IgG1 VH/VL pairs
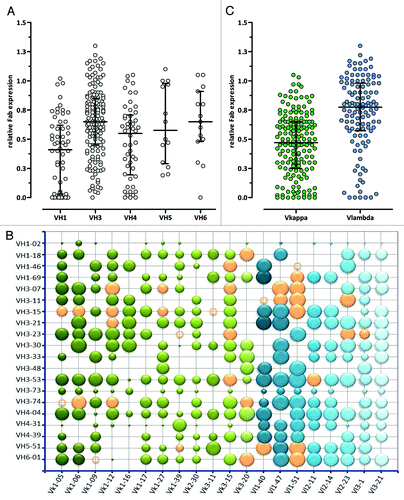
Of the theoretically possible 400 VH/VL combinations, 285 different VH/VL pairs were identified following a pooled cloning approach. They were analyzed for relative Fab expression in bacterial cell lysates by ELISA (), where the expression level of each VH/VL pair was determined relative to the expression of the reference Fab VH1–69/Vλ1–40. More than 70% of the tested Fab VH/VL pairs showed a relative expression of at least 0.4 of the reference Fab. VH3 containing VH/VL pairs had, on average, the highest relative Fab expression levels. The difference to VH/VL pairs belonging to other VH families, however, did not reach any statistical significance (). Of all tested VH/VL Fab pairs, VH1–2 and VH3–73 containing Fabs yielded consistently the lowest relative expression (), which correlated with the low relative Fab display rate observed for these frameworks as described below. Interestingly, Fabs with a Vκ framework showed a lower median relative expression than Vλ bearing Fab fragments (median 0.47 vs. 0.78, p < 0.0001, Mann-Whitney U-test) in the bacterial cell lysate screening ().
Figure 4. Relative expression levels of different VH/VL Fab pairs determined by ELISA. (A) VH, (B) VH/VL and (C) VL (Vκ: green, Vλ: blue) relative display rates are shown compared with an internal reference Fab VH/VL pair. In (A) and (C) the median with interquartile range is shown, in the bubble chart (B) the bubble area correlates with relative Fab expression. Orange bubbles and circles show the final selected 36 VH/VL pairs that are included in Ylanthia. The circles indicate VH/VL pairs that were not determined.
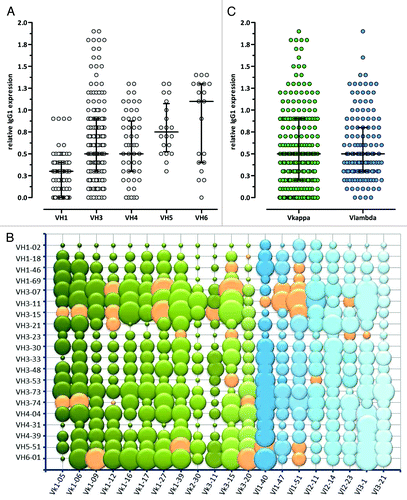
For human IgG1 expression, the 40 V region fragments were cloned into a IgG1 expression system encoding Ig heavy and Ig light chain on separate plasmids. In this way, all possible 400 VH/VL combinations could be assessed for small-scale IgG1 expression levels in HEK293 EBNA cells by ELISA (). Relative expression yields were calculated by dividing the signal of the respective VH/VL pair by the signal of a high level expressing reference IgG1/λ antibody (MOR03080).Citation49 Of the 400 tested VH/VL pairs, 209 (52%) showed an expression level equal to or above the level of the reference antibody with no major variance between VH families (). Surprisingly, all VH1–2 and VH3–23 containing IgG1s showed reduced expression levels compared with the reference antibody (). No significant difference between the median IgG1 expression levels of VH/Vκ and VH/Vλ frameworks (, median 0.5 vs. 0.5, p = 0.59, Mann–Whitney U-test), and no universal correlation between bacterial Fab and mammalian IgG1 expression levels was observed.
Figure 5. Relative expression levels of different VH/VL IgG1 pairs determined by ELISA. (A) VH, (B) VH/VL and (C) VL (Vκ: green, Vλ: blue) relative production yields are shown compared with a high expressing internal reference IgG1 antibody. In (A) and (C) the median with interquartile range is shown, in the bubble chart (B) the bubble area correlates with relative IgG1 expression. Orange bubbles and circles show the final selected 36 VH/VL pairs that are included in Ylanthia. The circles indicate VH/VL pairs that were not determined.
Relative display rates of Fab VH/VL pairs
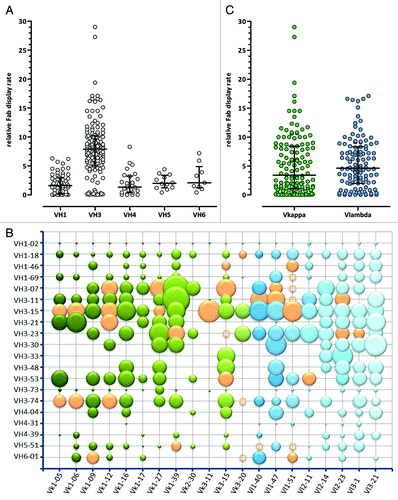
To ensure high display rates on phage, which is required for successful phage pannings, 243 VH/VL framework pairs were analyzed for relative Fab CysDisplay on phage by ELISA (). The relative display rate of VH3 containing VH/VL Fab pairs was significantly higher with an average of 8.0, compared with the relative display rates of VH1 (1.9), VH4 (2.1), VH5 (2.2) and VH6 (2.8) containing VH/VL pairs (only VH3 vs. VH1, VH3 vs. VH4, VH3 vs. VH5 and VH3 vs. VH6 gave significant differences, p < 0.05, one-way analysis of variance, Bonferroni’s multiple comparison test) (). Within the VH3 pairings, VH3–73 containing Fabs showed, on average, the lowest relative Fab phage display rate (0.2). Low Fab display and expression were also observed for VH1–2, VH4–31, VH4–39 and VH5–51 containing VH/VL pairs (). Between VH/Vκ and VH/Vλ pairs, no significant difference in relative Fab phage display rates was detected (median 4.4 vs. 4.6, p = 0.30, Mann–Whitney U-test) ().
Figure 6. Relative display rates of different VH/VL Fab pairs determined by ELISA. (A) VH, (B) VH/VL and (C) VL (Vκ: green, Vλ: blue) relative display rates are shown compared with an internal reference phage preparation. In (A) and (C) the median with interquartile range is shown, in the bubble diagramm (B) the bubble area correlates with relative Fab display. Orange bubbles and circles show the final selected 36 VH/VL pairs that are included in Ylanthia. The circles indicate VH/VL pairs that were not determined.
Thermal stability screening of Fab VH/VL pairs
The apparent melting temperature (Tm), a predictor for thermodynamic stability and resistance to unfolding, was assessed in the Fab format using bacterial cell lysates incubated at four different temperatures (0°C, 60°C, 70°C and 80°C) for a defined period of time. Integrity of the Fab molecules was assessed by ELISA using conformation-sensitive antibodies for detection. Denatured, aggregated or degraded Fab molecules were expected to show reduced signals. Of the 285 tested Fab VH/VL pairs, 215 (75%) showed a Tm of at least 60°C. In addition, our results suggest that the majority of VH/VL combinations comprising Vκ1–17 and Vκ2–30 are not stable at temperatures above 60°C, whereas most VH/VL pairs containing Vκ1–27 and Vκ3–15 were stable at 70°C (data not shown). Although this experiment was not suited to determine the exact Tm of the Fab molecules, this screening approach allowed rapid identification of VH/VL pairs sensitive to elevated temperatures.
In conclusion, the screening results showed that VH/VL framework pairs differ in their relative Fab display rate on phage, their Fab and IgG1 expression levels and thermal stability in vitro. This combined experimental data set was used as a basis for selecting 95 promising VH/VL pairs for further detailed characterization. The previous screening assays were useful in identifying VH/VL pairs containing instable Ig light chains, such as Vκ1–17 and Vκ2–30 or low expressing Ig heavy chains, e.g., VH1–2 and VH3–73. Such Ig heavy and Ig light chains were consequently excluded from all further evaluations.
Biophysical characterization of pre-selected Fab and IgG1 VH/VL pairs
On the basis of these screening results, 95 VH/VL pairs were selected for a more detailed analysis. This detailed examination of purified VH/VL pairs included the quantification of research-scale Fab and full-length human IgG1 expression levels, the analysis of aggregation propensity and apparent thermal stability, as well as a pH and physical stress testing as predictors for optimal antibody characteristics during manufacturing, formulation and process development.
Research-scale expression yields of Fab and IgG1 VH/VL pairs
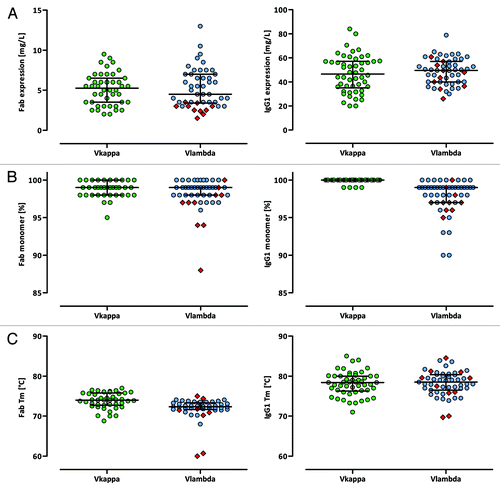
The synthetic antibody gene sequences within Ylanthia were codon-optimizedCitation27 for both E. coli (Fab) and mammalian expression (human IgG1) to obtain high Fab and IgG1 expression levels necessary for high-throughput screening platforms. The research-scale Fab expression yields of 91 VH/VL pairs measured after purification ranged from 1.5–13 mg/L with a median yield of 5.0 mg/L. In contrast to the bacterial cell lysate Fab expression screening of a larger set of random VH/VL pairs (see above), no major difference in Fab expression yields was observed between the selected Vκ and Vλ bearing VH/VL pairs. This likely reflects the bias introduced through the pre-selection of well-expressing VH/VL framework combinations (median 5.3 vs. 4.5, p = 1.00, Mann–Whitney U-test). All but one of the 91 Fab samples resulted in amounts greater than 2 mg/L, with only VH/ Vλ3–1 pairings showing relatively low expression levels (, left). VH/VL combinations in human IgG1 format showed moderate to high expression yields in HKB11 cells ranging from 20–80 mg/L with a median yield of 49.5 mg/L. Again, no significant difference in expression levels was detected between Vκ and Vλ bearing VH/VL IgG1 pairs (median 46.6 vs. 49.6, p = 0.5, Mann–Whitney U-test) (, right).
Figure 7. Biophysical features of human Fab and IgG1 VH/VL pairings. (A) Production yields of purified Fab (left panel) and human IgG1 (right panel) molecules after purification as determined by UV-spectrophotometry. (B) Monomer contents of purified Fab (left panel) and human IgG1 (right panel) molecules as determined by SEC. (C) Apparent melting temperatures of purified Fab (left panel) and human IgG1 (right panel) molecules as determined by DSF measurements. In each panel the median with interquartile range is indicated. VH/Vλ3–1 pairs are highlighted as red squares.
Aggregation propensities of purified Fab and IgG1 VH/VL pairs
The presence of multimeric and aggregated Fab and human IgG1 molecules following purification was evaluated by high-performance analytical size-exclusion chromatography (HP-SEC). The monomeric portions of the purified VH/VL pairs ranged between 88% and 100% with a median monomer portion of 99% each for VH/Vκ and VH/Vλ Fab fragments and VH/Vλ IgG1 molecules. VH/Vκ IgG1 antibodies showed a median monomer content of 100%. Compared with Vκ combinations, Vλ bearing Fab and IgG1 VH/VL frameworks seem to be more prone to aggregation (Fab: p = 0.03, Mann–Whitney U-test, , left; IgG1: p < 0.0001, Mann–Whitney U-test, , right). The vast majority (96%), however, showed monomeric portions above 95%, indicating a very low aggregation tendency. Furthermore, electrophoresis-based analyses under denaturing reducing conditions demonstrated purities above 92% (data not shown) with no precipitations occurring during the purification process.
Melting temperatures (Tm) of purified Fab and IgG1 VH/VL pairs
The apparent Tms of the purified Fab molecules determined by differential scanning fluorometry (DSF)Citation50-Citation52 ranged from 50.9°C (VH3–07/Vλ3–1) to 77°C (VH3–15/Vκ3–15) with a median Tm of 74.0°C for Vκ and 72.4°C for Vλ containing frameworks (p < 0.0001, Mann–Whitney U-test, , left). In addition, the second transition Tms of the IgG1 antibodies, which represents the stability of the Fab portion, ranged from 69.7°C (VH3–07/Vλ3–1) to 85.0°C (VH3–23/Vκ3–15) with a median Tm of 78.4°C and 78.5°C for Vκ and Vλ IgG1 combinations, respectively (p = 0.68, Mann–Whitney U-test, , right). The human IgG1 Fc domain shows a transition between 68.4 and 70.6°C (data not shown). In cases with only one single transition, the Tm of the Fab portion coincides with the melting temperature of the Fc part. Within the VH/VL IgG1 pairs of Ylanthia finally selected, all Fab domains showed a Tm equal or higher to 70°C.
The Tm values of the Fab and IgG1 molecules did not change significantly after acidic stress exposure, indicating that the molecular structures remain unaffected by pH-stress conditions or refold efficiently after stress relief. We conclude that the selected Fab and IgG1 VH/VL framework pairs are very stable regarding temperature unfolding after acid exposure.
Molecular size measurements of purified IgG1 VH/VL pairs
To study the molecular size of the purified human IgG1 antibodies, the hydrodynamic radius and polydispersity were evaluated for each sample by dynamic light scattering (DLS). Out of 89 tested IgG1 VH/VL framework pairs, 80 VH/VL combinations showed a hydrodynamic radius below or equal to 6 nm and were therefore considered to exist as monomeric IgG1. Seven VH/VL pairs (VH3–15/Vκ1–09, VH1–46/Vλ3–1, VH1–46/Vλ3–21, VH1–69/Vλ1–40, VH1–69/Vλ3–1, VH3–21/Vλ3–1, VH3–30/Vλ3–1) showed a radius between 7 and 14 nm and two framework pairings (VH6–1/Vλ1–47 and VH6–1/Vλ3–1) exhibited a radius higher than 20 nm, indicating the presence of higher molecular weight aggregates (). Interestingly, the Vλ3–1 framework is present prevalently within these VH/VL pairs.
Figure 8. Polydispersity of purified human IgG1 antibodies. The hydrodynamic radius of purified VH/VL IgG1 molecules was determined by DLS experiments. VH/Vλ3–1 pairs are highlighted as red squares.
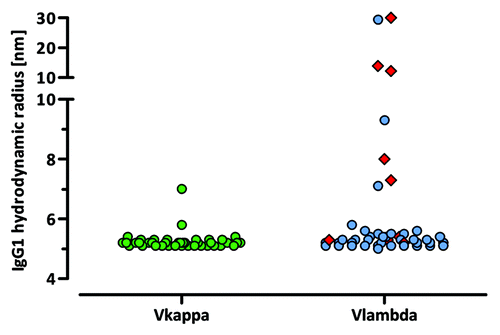
For the majority of VH/VL pairings, the hydrodynamic radius remained unaltered after acidic treatment (data not shown). Six of the 9 antibodies that had a radius higher than 7 nm before treatment displayed a further increase in their hydrodynamic radius after acidification, indicating gradual aggregation.
The initial polydispersity values of 85 tested VH/VL pairs ranged from 2 to 40% with 11 antibodies exceeding the 15% cut-off, indicative of a potential IgG1 heterogeneity. For 6 out of the 74 initially homogenous antibodies, acid stress treatment led to heterogeneous particle size distributions (i.e., polydispersity > 15%). Such an increase in sample heterogeneity can be explained by factors such as dimer formation or conformational changes of the respective IgG1 molecules.
Turbidity of purified IgG1 VH/VL pairs
UV-spectrometry measurements at 320 nm were performed to determine the turbidity of IgG1 samples before and at different time points (10 min and 100 min) after acidification and subsequent neutralization. Except for three VH/VL pairs (VH1–46/Vλ3–1, VH6–1/Vλ1–47, VH6–1/Vλ3–1) that already showed a relatively high initial extinction, all other tested VH/VL pairs showed no or only a moderate increase in turbidity upon low pH treatment, indicating a relative resistance toward aggregate formation. The morphology of visible and sub-visible aggregates in solution was further assessed by a semi-quantitative particle staining assay (data not shown).
Library composition, CDR3 design and library cloning
Based on the above analyses, 36 VH/VL framework pairs with good expression and display properties and superior biophysical features were selected as frameworks for the Ylanthia library. An overview of the biophysical data of the finally selected 36 VH/VL framework pairs is given in and .
Table 2A. Igk VH/VL pairings that constitute the final Ylanthia antibody library
Table 2B. Ig λ VH/VL pairings that constitute the final Ylanthia antibody library
The 36 selected VH/VL framework pairs comprise 12 VH, 9 Vκ and 6 Vλ gene segments. As the Ig light chain CDR-L1 and CDR-L2s are for the most part devoid of potential PTM hot spots, the germline sequences were utilized. In contrast, the 12 VH gene segments contain CDR sequences that are fully germline, as well as CDR-H1s and CDR-H2s that have been diversified to exclude potential PTM sites.
In contrast to the HuCAL conceptCitation19 with its major Fab libraries HuCAL GOLDCitation21 and HuCAL PLATINUM,Citation20 the Ylanthia library is neither based on random combinations of VH and VL domains nor modularity of the 6 CDRs. In Ylanthia, for the CDR-L3 and CDR-H3 diversity, the natural length variations present in the human antibody repertoire were closely mimicked. A Kabat CDR-L3 length of 9 amino acids was implemented in the design for the majority of the Vκ regions. For Vκ3 and for all Vλ regions CDR-L3s, the lengths of 9 and 10 amino acids, and the lengths of 9 to 11 amino acids, respectively, were realized. The composition of each CDR-L3 cassette was designed according to the corresponding VL gene segment region that naturally contributes to CDR-L3.
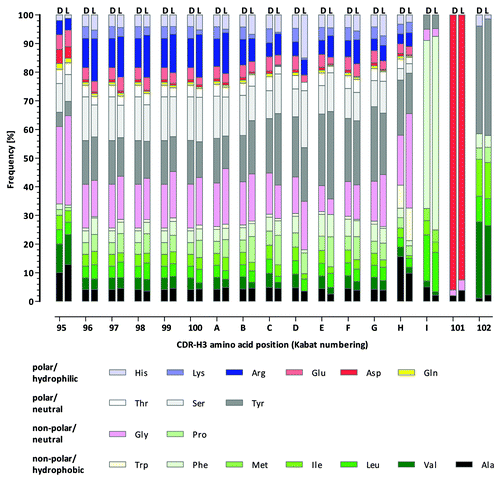
Natural CDR-H3 diversity is mainly generated by VDJ recombination of different VH, D and JH gene segments that result in CDR-H3 sequence length variations from a few to more than 30 amino acids.Citation53 In Ylanthia, 12 different CDR-H3 lengths ranging from 6 to 17 amino acids were implemented. These CDR-H3 lengths cover about 80% of naturally occurring CDR-H3 sequences as described by Zemlin and coworkers.Citation53 The CDR-H3 length and amino acid composition design was identical for all VH gene segments used. It has been observed that JH4 is used predominantly in rearranged human antibodies and that the incorporation of the longest JH6 gene segment gains importance with increasing CDR-H3 lengths, resulting in subtle differences in positional amino acids diversities.Citation53 Therefore, the realized amino acid composition in the CDR-H3 is based on naturally rearranged Ig sequences using JH4, where the composition of the lengths 12 to 17 amino acids has an additional JH6-based design closely reflecting the CDR-H3 length-dependent amino acid frequencies in the human antibody repertoire.
To abolish or reduce the occurrence of potential critical PTM sites in the CDR-L3 and CDR-H3, the amino acids asparagine (Asn, N), aspartic acid (Asp, D) and methionine (Met, M) were decreased or completely omitted, as, for example, in the case of Asn in CDR-H3 sequences. The overall CDR-H3 design is schematically illustrated in .
Library size, sequence correctness and diversity
The theoretical diversities for the Vκ region and Vλ region CDR-L3 range from 3.8E+06 to 2.5E+9. For the smaller Vκ CDR-L3 libraries, we aimed for a minimum library size of 5-fold above the theoretical diversity. For the more diverse Vλ CDR-L3 libraries, the size was restricted to approximately 5.0E+08 clones. The final sizes for each of the VH/VL pairs were achieved during the CDR-H3 cloning step and ranged from 4.6E+08 to 4.4E+09 members, which results in an overall library size of 1.3E+11 clones.
The quality of the library on a sequence level was assessed at three stages during the cloning procedure. This included assessment of sequence correctness as defined by correct reading frames and agreement of the amino acid distributions at the diversified CDR-L3 and CDR-H3 positions with the library design. The first quality control (QC) was performed directly after the synthesis of the CDR-L3 and CDR-H3 library cassettes, prior to their insertion into the respective vector backbones. A second QC step confirmed the VL sequences after the first cloning step (i.e., the insertion of the CDR-L3 cassettes). VL sequences were checked again in a third QC together with VH sequences after the final cloning step (i.e., the insertion of the CDR-H3 cassettes). Each QC step was performed by sequencing at least 30 clones of each of the respective V regions. The average sequence correctness of the intermediate Ig light chain sub-libraries was 85% for all Ig lambda and 86% for all Ig kappa sub-libraries. After the insertion of the CDR-H3 cassettes in the second cloning step, the CDR-L3 sequence correctness remained unchanged and the average CDR-H3 correctness alone of all sub-libraries was 90% resulting in a combined CDR-L3 and CDR-H3 sequence correctness of at least 82% for each sub-library.
The initial CDR-L3 and CDR-H3 QC also included the comparison of the amino acid composition of each variable position synthesized with that of the library design. As shown in , no major deviations between the observed composition in the unselected library and the library design were observed for the CDR-H3 lengths 6 to 17 for roughly 4,000 analyzed sequences. In addition, the CDR-H3 length variability observed in the library prior to antigen selection very closely mirrored the expected length distribution ().
Figure 10. CDR-H3 length distribution in Ylanthia. CDR-H3 lengths according to the library design (white bars) and as determined after small QC (gray bars, analysis of ~3,200 sequences) and after 454 sequencing (black bars, analysis of > 152,000 sequences).
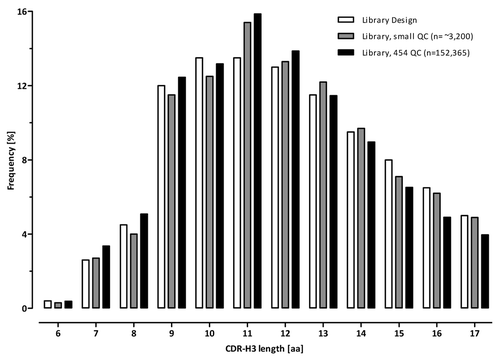
To further confirm the sequence diversity and to check the VH, Vκ and Vλ framework distribution, an additional in-depth analysis of the entire Ylanthia library was performed by high throughput 454 sequencing using the Roche GS Junior System. The sequencing data were evaluated using proprietary software tailored for analysis of large sequencing data sets. For CDR-H3 length analysis, a total of 160,521 high quality reads (> 300 bp) were generated, of which 152,365 could be assigned to a corresponding length-specific group. As shown in , the CDR-H3 length distribution was congruent with the profile obtained in the small scale QC described above, with a slight reduction of the longest length variants. For codon-based CDR-H3 redundancy analysis, the occurrence of sequence replicates was compiled from the same data set (> 152,000 sequences). The complete library was found to be composed of 94% unique nonredundant antibody CDR-H3 sequences. The remaining ~6% of the library was found as duplicate CDR-H3 sequences (5.4%) and to a very low percentage as tri- and quadruplicates, which together accounted for 0.38% (). Importantly, CDR-H3 sequence diversity as defined by the ratio of the number of different sequences divided by the total number of sequences was greater than 97%.
Figure 11. Codon-based sequence redundancy in Ylanthia as determined by 454 sequencing. VH region sequences were amplified using pan-selective primers recognizing all Ig heavy chain sequences. From more than 152,000 analyzed VH sequences 94% were unique in their CDR-H3 with less than 6% of sequences found in duplicates, indicating high sequence diversity.
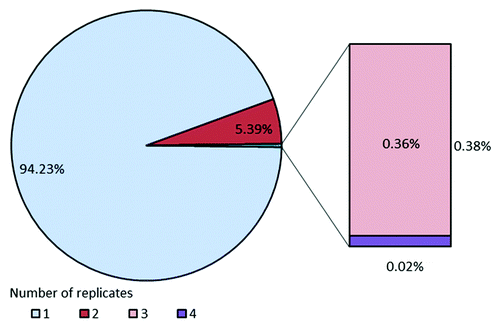
Of note, sequence analysis of about 2,000 clones from the initial “Slonomics” CDR-H3 library and of more than 4,000 clones of the final library by Sanger sequencing has not revealed any detectable redundancy (data not shown). This observation is leading to the suggestion that the major portion of the observed replicates originates most likely from an amplification bias introduced during the 454 workflow.
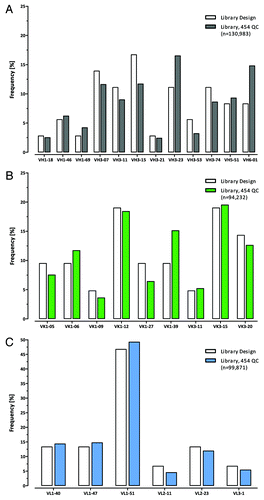
For framework distribution analysis the frequencies of each VH, Vκ and Vλ framework were compiled from sequences (> 90bp) that could be unambiguously assigned to specific frameworks. For VH, Vκ and Vλ 130,983, 94,232 and 99,871 sequences, respectively were evaluated. The VH (), Vκ () and Vλ () framework frequencies were found to the portions as expected by the design indicating an unbiased cloning performance during the construction of the Ylanthia library.
Figure 12. VH, Vκ and Vλ framework distribution in Ylanthia as determined by 454 sequencing. By analyzing more than 325,000 sequences in total, the observed distribution of the (A) VH (gray bars), (B) Vκ (green bars) and (C) Vλ (blue bars) frameworks closely represented the expected frequencies by design (white bars) in the library.
Antibody selections
Phage preparations for each sub-library were performed as outlined in the materials and methods section and phage titers and relative Fab display rates were determined by UV-spectroscopy and ELISA, respectively (data not shown). Antibody phage selections (‘pannings’) were performed according to standard protocols.Citation54,Citation55 The following different protein antigens were used to perform phage selections: recombinant human (rh) ErbB4, rhFZD4/Fc, rhTNFalpha, M-CSF, eGFP, a human IgG1 antibody (with the goal to generate anti-idiotypic antibodies) and three other antigens. After three rounds of pannings, the enriched Fab encoding sequences were subcloned as pools into the pYBex10 Fab expression vector. Antigen-specific Fabs were identified by ELISA, where a positive hit was defined by binding to its specific target with a signal at least 5-fold above the assay background.Citation54,Citation55 Overall, 2,900 to 5,200 clones were screened per target antigen with primary hit rates ranging from 1% to 49%. Sequencing of up to 785 positive hits revealed 7 to 284 CDR-H3 unique antibodies per antigen, although the total diversity of selected antibodies will most likely be much higher because only a minor fraction of the clonal panning output was screened and sequenced. Notably, a low hit rate was only observed with challenging targets, e.g., anti-idiotype selection, due to the relative scarcity of unique epitopes on the target antigen (binding interface of human IgG1 antibody). So far, antibodies from 27 distinct VH/VL combinations (out of 36 VH/VL pairings) were identified. Different target antigens thereby favor different VH/VL pairings (data not shown), arguing for increased framework diversity representing an advantage over current in vitro technologies for antibody discovery on complex targets. Monovalent affinities were determined by surface plasmon resonance measurements (Biacore/ GE Healthcare) only for a small subset of antibodies. KD values as low as 700 pM (kon 2.4E+05 M−1s−1, koff 1.7E-04 sec−1]) against a protein antigen were found confirming that high affinity antibodies can be isolated directly from the library (Fig. S1). In the subset of 24 analyzed Fab molecules, the KD values ranged between 700 pM and 190 nM with kon rates between 2.9E+04 M−1s−1 and 4.6E+06 M−1s−1 and koff rates between 8.1E-05 sec−1 and 1.7E-01 sec−1 (Table S1).
Biophysical properties of selected Ylanthia IgG1s
For each of the 9 individual panning experiments, up to 30 Fab hits were converted into human IgG1 format, resulting in a total of 312 IgG1 antibodies that were produced in research-scale and characterized according to their biophysical properties. The calculated pI values of the IgG1 antibodies ranged from 7.6 to 9.7 with an average pI of 9.2, with no significant differences between Vκ and Vλ IgG1s (). Purified human IgG1 yields in a transient HKB11 mammalian expression system ranged from 1 mg/L to more than 60 mg/L with an average expression level of 23 mg/L. Interestingly, Vλ IgG1s, with an average of 28 mg/L, showed significantly higher expression levels than Vκ IgG1s, with an average of 17 mg/L (p < 0.0001, t-test) (). The aggregation propensity, as determined by HP-SEC experiments, was very low for the majority of binders. Of 259 tested IgG1 antibodies, 232 (89.6%) showed a monomer content equal to or higher than 90% and 196 (75.7%) had a monomer portion greater than 95% (). As an additional indicator for protein stability, the IgG1 melting temperatures were determined by DSF experiments. Of the tested IgGs, 233 (89.6%) had a melting temperature equal to or higher than the temperature corresponding to the unfolding of the Fc CH2 domain that occurs at about 68°C. The median Tm value of all Vκ IgG1 antibodies was significantly higher compared with the Vλ bearing IgG1s (70.9°C vs. 77.9°C, p < 0.0001, Mann–Whitney U-test) as shown in .
Figure 13. Biophysical features of selected Ylanthia IgG1 antibodies. (A) pI values, (B) production yields after purification, (C) monomer contents and (D) apparent melting temperatures of purified human Vκ (green) and Vλ (blue) IgG1 antibodies that were selected in various panning campaigns. In each panel the median with interquartile range is indicated. Grey shaded regions indicate the respective parameters preferred range that is thought to be optimal for antibody developability.
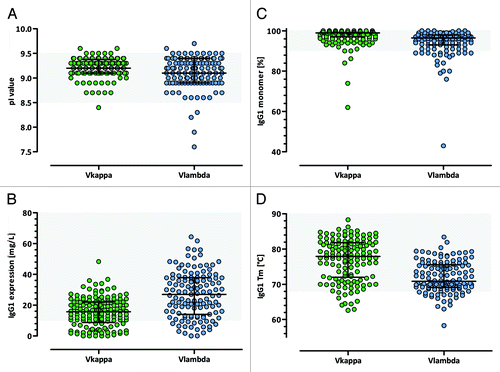
In conclusion, the majority of the selected IgG1 antibodies showed optimal biophysical characteristics that were comparable to the features initially determined for the corresponding VH/VL framework pairs.
Discussion
In vitro display methods using recombinant libraries have been firmly established in the field of human antibody generation and are highly valued for their ability to deliver high affinity antibodies with tailor-made specificity. As large numbers of therapeutic antibody candidates have progressed from discovery through development, a great deal of knowledge has accumulated regarding the desired molecular properties of antibody drug candidates. Biophysical properties, such as thermodynamic stability and folding efficiency influence the solubility and aggregation behavior of antibodies. These features, therefore, are important factors in protein expression, manufacturing and formulation. Such features can also affect both pharmacokinetics and immunogenicity, altogether having important consequences for the therapeutic potential and the ease of use of an antibody.Citation56 Engineering to optimize the biophysical properties of any given candidate has therefore become an important aspect of antibody discovery. Rather than having to perform time and cost-intensive optimization, it would, however, be preferable to isolate antibodies that fulfill such desired criteria at the start. With this goal in mind, an antibody library that is optimized for “processability” has been generated. Presented here are the design, construction and application of Ylanthia, a fully synthetic human Fab antibody phage display library built on 36 fixed germline VH/VL framework pairs selected for their favorable biophysical properties.
Our vision was to identify germline VH/VL framework pairs with optimal biophysical properties from the approximately 3,200 theoretical functional VH/VL combinations of the human Ig repertoire. We began with the hypothesis that the interplay between Ig heavy and Ig light chain is important and that a co-selection of fixed pairs may enable the identification of frameworks with superior characteristics. In a first step, 20 diverse segments were selected for both VH and VL based, in part, on their frequency in naturalCitation30-Citation37 and engineered antibody repertoires,Citation41 assuming that preferred germline usage correlates with antibody expression and stability.Citation57 In a second step, the selected variable domains were recombined with defined CDR3sCitation19,Citation47,Citation48 and the majority of the resulting 400 VH/VL pairs were screened to assess display and expression properties both as Fab and IgG1. The rationale to look at Fab and IgG1 expression was that low expression levels often reflect sub-optimal folding properties.Citation58 High Fab display rates in turn are essential for successful phage selections because they maximize Fab–antigen interactions during phage selections. From this screening, 95 VH/VL pairs were selected for further in-depth characterization of expression and other biophysical characteristics in both Fab and IgG1 formats.
During manufacturing processes, therapeutic antibodies are subjected to a range of conditions that are likely to perturb their colloidal solubility and native conformation. We selected assays for further characterization of our framework VH/VL pairs to simulate those stress conditions and select for properties that ensure optimal performance. As an example, chemical stress is generated during virus inactivation by acid treatment that was meant to denature viral capsid proteins.Citation59,Citation60 Low pH, however, destabilizes the antibody such that the processed protein eventually unfolds and denatures. To exclude VH/VL pairs that are sensitive to low pH, DSF and DLS experiments were performed to determine the apparent Tm and the hydrodynamic radius of purified VH/VL pairs under different pH conditions. The determination of the melting point provides a useful measure for protein stability and conformational integrity. In general, low Tm values indicate low stability of the VH/VL protein pair that might lead to higher aggregate levels in production batches, demanding increased efforts to stabilize the respective protein in appropriate buffer formulations.Citation61 To further predict physical stability and aggregation propensities of the VH/VL pairs during formulation and storage, the oligomer status in solution was analyzed by extinction measurements and particle staining. It is worth noting that all engineered Fab molecules are devoid of an intermolecular disulfide bond between Ig heavy and Ig light chain that would otherwise stabilize the molecule.
Through this extensive characterization, 36 fixed VH/VL framework pairs with superior biophysical and expression properties were selected, excluding VH/VL pairs with sub-optimal performance. These 36 VH/VL pairs comprise 12 different VH gene segments and 15 distinct VL gene segments (9 Vκ, 6 Vλ). Together they cover most of the structural space of naturally occurring canonical CDRs and mirror closely the occurrence of human V gene families in natural antibody repertoires. The only exceptions are frameworks that do not display efficiently on phage, such as VH2, VH4 and Vκ4 as these were completely omitted. Previous experience, for example with scFv libraries,Citation19 HuCAL GOLDCitation21 and the CAT2.0 library,Citation41 has shown that VH4 gene sequences were rarely selected and it is believed that this might be at least in part due to low display rates (unpublished observations; data not shown) and unfavorable biophysical characteristics of this framework.Citation19,Citation48
In contrast to earlier work that failed to reveal any evidence for preferences in natural pairing of particular VH and VL gene families,Citation39,Citation62 it was recently suggested that the human VH1 family shows a strong pairing preference with the Vκ3 family.Citation57 Interestingly, two of the four VH1 family members within Ylanthia pair with Vκ3 members (i.e., VH1–18/Vκ3–20, VH1–46/Vκ3–15), indicating that this naturally occurring prevalence is reflected in favorable biophysical properties. Our observation of an increased tendency to aggregate for Vλ bearing VH/VL framework pairs (especially the Vλ3–1 framework) might be explained by their particularly low pI values or it could be an intrinsic feature of the primary sequence of the Vλ proteins. Isolated Vλ consensus sequences have been described to be relatively unstable by themselves with a tendency to dimerize, although scFv fragments containing Vλ domains were shown to possess partly very high stability due to a strong interaction between the VH and Vλ domains.Citation48 Furthermore, VH/VL pairings containing Vλ1 family members overall showed the lowest Tms, whereas VH/VL pairs with Vκ domains showed a tendency toward higher Tms. This might again be influenced by the generic amino acid composition of Vλ sequences, as well as the sequence and structural differences of Vλ and Vκ CDR-L3.Citation44,Citation63
The fully synthetic character of Ylanthia allowed for complete control of both nucleotide and amino acid content and distribution and the convenient realization of CDR-Hs that cover germline sequences as well as sequences without potential PTMs. The use of VH sequences devoid of PTMs in their CDR-H1 and CDR-H2 make the subsequent removal of such motifs unnecessary; however, they harbor the risk of being potentially immunogenic in humans. To keep the risk for immunogenicity (i.e., the generation of T cell epitopes) at a minimum, the required amino acid exchanges were performed in a controlled way. All Ig sequences were also adapted for optimal codon usage in E. coli and in mammalian cells providing adequate conditions for convenient production of Fab as well as human IgG1.Citation27
The major challenge in cloning large antibody libraries is producing a large number of functional members. In Ylanthia, CDR-H1 and -H2 are diversified, but only CDR-L3 and CDR-H3s were randomized, as these regions have the strongest influence on antigen-binding and affinity. To ensure high quality and sequence correctness the CDR-L3 cassettes were synthesized using preassembled trinucleotides (TRIMs)Citation28 and the CDR-H3 cassettes were synthesized through the Slonomics technology.Citation29 While in nature, CDR-H3 length varies from a few amino acids to more than 30 residues,Citation53 only CDR-H3 lengths of 6 to 17 amino acids were implemented in Ylanthia, as these lengths represent the majority of naturally occurring CDR-H3 sequences. In addition, potential critical PTM sites have been omitted or reduced in the CDR3s to prevent later antibody heterogeneity, instability or even loss of function through post translational modifications.Citation64 Furthermore, the diversified CDR-L3 and CDR-H3 sequences were designed to obtain pI values ranging between 7.5 to 9 for the full-length antibody. This should maximize solubility of the protein in the neutral or slightly acidic formulation buffers routinely used. For antibodies with pI values greater than 9 rapid clearance from the blood and decreased half-life in vivoCitation65,Citation66 have been described.
For the generation of Ylanthia, the diversified CDR-L3 and CDR-H3 cassettes were cloned consecutively into each of the 36 Ylanthia VH/VL pairs, without the use of an in-frame selection strategy, resulting in a total antibody library size of more than 1.3E+11 individual members. Thus, Ylanthia is 3-fold larger than the previously described HuCAL PLATINUM libraryCitation20 and equals the human PCR-derived CAT2.0 libraryCitation41 in size. To our knowledge, the quality of Ylanthia, with ~85% accurate clones is among the highest reported to date for recombinant antibody libraries. High-throughput 454 sequencing allowed for a precise and unbiased control of the library compositionCitation43 and comparison to the initial design. The degree of redundant CDR-H3 sequences, as revealed by 454 sequencing, is in line with the CDR-H3 redundancy observed after next generation sequencing of other antibody libraries with fully synthetic CDR3s. For example, a scFv library generated by Nicolas Fischer and his colleagues exhibited 98.5% unique sequencesCitation67 and the HuCAL Platinum libraryCitation20 where 454 sequence analysis of several Ig heavy chain sub-libraries was performed resulted in 98% unique CDR-H3 sequences (Josef Prassler, personal communication). Interestingly, CDR-H3s amplified from naïve human donors or from mice seem to exhibit much lower diversity with only 24% to 31% unique CDR-H3 sequences as described by Venet et al.Citation68
To maintain the convenient selection process of high-affinity antibodies established for HuCAL GOLDCitation21 and PLATINUM,Citation20 the Fab fragments are displayed by CysDisplay through disulfide linkage to the pIII protein on the phage. More than 300 IgGs against recombinant proteins including soluble peptides and receptors have been identified in various pilot experiments with Ylanthia. Thus, a large amount of data has already been acquired to assess the expression characteristics and biophysical properties of antibodies derived from Ylanthia and to underscore its potential to select for antibodies with preferred “processability” characteristics.
Among the selected binders, Vλ IgG1 antibodies yielded higher expression levels compared with Vκ bearing VH/VL IgG1 pairs. Interestingly, this difference observed following selection on a target had not been present in the initially tested 100 VH/VL pairs, indicating a potential influence of the selected CDR3s. Regarding monomer content, the behavior of the 36 VH/VL framework pairs (not more than 2% aggregates) is largely preserved in the VH/Vκ IgG1 output from phage pannings. As already discussed above for VH/Vλ pairs containing Vλ3–1, they seem to be more prone to aggregation. An explanation for this increased aggregation tendency may be found in the relatively flexible Vλ CDR-L3 compared with more rigid Vκ CDR-L3 sequence compositions.Citation44,Citation63,Citation69
Nevertheless, the majority of the antigen-selected antibody molecules closely preserved the biophysical features that were characteristic of the corresponding “parental” VH/VL framework pair. Slight biophysical deviations were observed only in a minority of selected binders and thus have to be attributed to the influence of the particular CDR-L3 and CDR-H3 sequences. While such effects remain difficult to circumvent without abating overall CDR3 diversity, a very good balance between biophysically optimal and diverse VH/VL pairs has been obtained after phage selections.
The Ylanthia library is thus based on a novel concept that incorporates desirable antibody characteristics in its design through selection of optimal framework pairs and careful design of the CDRs. It represents a new platform that generates fully human antibody candidates with optimized biophysical properties to ensure straight-forward and fast antibody processing and development. In many cases Ylanthia antibodies may not require further engineering and thus may save valuable time in the discovery process.
Materials and Methods
Evaluation of the human antibody repertoire and CDR3 analysis
To identify the most prevalent VH and VL gene segment pairs in the human Ig repertoire, various single B cell PCR studies from literature and from sampling of single human B cells in-house were evaluated.Citation30-Citation35 Single cell sorts, Ig gene amplification and sequencing were basically performed as previously described.Citation5
For CDR3 analysis, rearranged human antibody sequences were obtained from single B cell PCR experiments and from various Ig V-gene sequence databases such as V-BASECitation70 and IgBLAST.Citation71 The amino acid sequences were imported into Microsoft Excel and CDR3 regions were analyzed by using custom-made Visual Basic utilities as described before.Citation19-Citation21,Citation72
Gene synthesis and assembly
V regions with flanking sites for restriction enzymes were synthesized by GeneArt with CDR-H3 (WGGDGFYAMDY) and kappa CDR-L3 (QQHYTTPPT) of the hu4D5–8 antibodyCitation47 and lambda-like CDR-L3 (QSYDSSLSGVV) sequencesCitation48 and the JH4, Jκ1 and Jλ2/3 germline protein sequences, respectively.Citation73-Citation75 Codon usage optimization with respect to E. coli expression (avoiding rare human codons) and removal of potential cis-acting sequence motifs such as RNA instability motifs and cryptic splice donor and acceptor sites were performed by GeneArt.Citation27
The synthesized Ig V region heavy chain genes covered the sequences from the first unique 5′ restriction site for NheI located in the modified phoA leader sequence to the unique 3′ restriction sites for XhoI and SalI located at the V to C region border. Upstream from the CDR-H3, a restriction site for BssHII was incorporated to enable subsequent CDR-H3 library insertion. The Ig Vκ and Vλ region light chain gene sequences ranged from the first unique 5′ restriction site for NdeI located in the modified ompA leader sequence to the 3′ restriction sites for Acc65I/KpnI in framework region (FWR) 4. Upstream from the CDR-L3, a restriction site for BbsI was incorporated to enable subsequent CDR-L3 library insertion.
Biophysical screening of Fab VH/VL pairs
The 20 VH region constructs were pool cloned together with 12 Vκ region or 8 Vλ region constructs into the tricistronic CysDisplay vector pYPdis10 and the bacterial Fab expression vector pJPx1_FH.
First, pools containing the Ig variable heavy chain regions (VH3–07, VH3–11, VH3–15, VH3–21, VH3–23, VH3–30, VH3–33, VH3–48, VH3–53, VH3–73, VH3–74, VH1–02, VH1–18, VH1–46, VH1–69, VH4–04, VH4–31, VH4–39, VH5–51, VH6–01) were inserted via the restriction sites for NheI and SalI, followed by insertion of the Vκ (Vκ1–05, Vκ1–06, Vκ1–09, Vκ1–12, Vκ1–16, Vκ1–17, Vκ1–27, Vκ1–39, Vκ3–20 Vκ3–11, Vκ3–15 and Vκ3–20) and Vλ (Vλ1–40, Vλ1–47, Vλ1–51, Vλ2–11, Vλ2–14, Vλ2–23, Vλ3–1 and Vλ3–21) pool with NdeI and Acc65I. For each step the ligation mixtures were electroporated into E.coli TOP10F’ (Invitrogen).
From the resulting pYPdis10 and pJPx1_FH constructs, polyclonal DNA was prepared and transformed into electro-competent E. coli TOP10F′ (Invitrogen) for subsequent Fab display on filamentous M13 phage or TG1F− for bacterial Fab expression. Each transformed pYPdis10 and pJPx1_FH pool was plated on LB/Cam/Gluc plates and clones were picked into 384-well plates and stored in 15% glycerol (“master plates”).
For identification of the respective VH/VL pair, clones were sequenced with Y_Fab_VH_for (5′-GATAAGCATGCGTAGGAGAAA-3′), Y_Fab_dis_VL_for (5-'TGTTGCCACCTTTATGTATG-3′) or Y_Fab_Bex_VL_for (5′-TGGAATTGTGAGCGGATAAC-3′).
Small-scale phage preparations
Antibody phage preparations comprising the majority of the 400 VH/VL pairs were produced in small scale using 96-well plates. In brief, random clones from the master plates of the 400 VH/VL pairs cloned in display vector pYPdis10 were inoculated in 2× YT/Cam/1%Gluc and incubated while shaking at 400 rpm until an OD600 nm of 0.5 was reached. The cells were then infected with Hyperphage (PROGEN), centrifuged and re-suspended in 2× YT medium with 34 µg/ml chloramphenicol, 50 μg/ml kanamycin and 0.25 mM isopropyl-β-D-thiogalactopyranoside (2× YT/Cam/Kan/IPTG) and further incubated at 22°C for at least 14 h for phage production.
Relative Fab phage display rate ELISA
Fab display on M13 phage was determined by ELISA using an anti-M13 antibody (Amersham) to capture the phage via the major coat protein pVIII and using an anti-Fd antibody (The Binding Site) that binds to the displayed Fab. Appropriate dilutions of the phage supernatants and reference samples were detected with an anti-M13 peroxidase conjugate (Amersham) and QuantaBlu™ (Pierce). Calculations of relative display rates for each sample were performed by dividing the anti-Fd titer by the titer of anti-M13. Titers were obtained from calibration curves of reference phage preparations.
Relative Fab expression ELISA
Random clones from the 400 VH/VL pairs pool cloned in pJPx1 were inoculated in 2× YT/Cam/0.1% Gluc and incubated for at least 6 h at 30°C while shaking. Fab expression was induced by adding IPTG to a final concentration of 0.5 mM and incubated overnight at 22°C and with shaking. E. coli cells were lysed by adding 2× BBS/EDTA/lysozyme buffer (24.7 mg/ml boric acid, 18.7 mg/ml NaCl, 1.48 mg/ml EDTA, 2.5 mg/ml lysozyme, adjusted to pH 8.0 with 10 M NaOH) and subsequently blocked with 12.5% milk powder, 0.05% Tween® in PBS. Fab expression was determined by ELISA using an anti-human Fd capture antibody (The Binding Site) and an AP-labeled anti-FlagM2 (Sigma) detection antibody with AttoPhos (Roche). Relative Fab expression levels were calculated by dividing the signal of the respective Fab VH/VL pair through the signal of the reference Fab VH1–69/Vλ1–40 pair expressed with pMORPHx11.Citation20
Fab temperature stability ELISA
Appropriate dilutions of bacterial lysates were exposed to different temperatures (0°C, 60°C, 70°C and 80°C) for 45 min. Intact Fab molecules were detected by ELISA using an anti-6×His capture antibody (R&D Systems) and AP-labeled anti-human Ig kappa or anti-human Ig lambda (Sigma) detection antibodies with AttoPhos (Roche).
Biophysical screening of IgG1 VH/VL pairs
For generation of the 400 VH/VL IgG1 germline pairs, the 20 VH region fragments were sub-cloned via NheI and XhoI into the pYMex20_h_IgG1f vector and the 12 Vκ and 8 Vλ region fragments via NdeI and Acc65I into pYMex20_h_Igkappa and pYMex20_h_Iglambda, respectively. For small scale IgG1 expression, HEK293 EBNA cells were grown under standard conditions (DMEM/10% FCS, 37°C, 5% CO2) in 96-well plates until 80–90% confluence and then co-transfected by Lipofectamine (Invitrogen) with equimolar amounts of the corresponding pYMex20 Ig heavy and Ig light chain expression plasmids. Forty-eight hours after transfection cell culture supernatants were harvested.
Relative IgG1 expression ELISA
IgG1 expression in cell culture supernatants was determined by sandwich ELISA using a mouse anti-human IgG (clone R10Z8E9)Citation76 capture antibody and the same antibody as biotin conjugate for detection with streptavidin-AP and AttoPhos (Roche). Relative IgG1 expression levels were calculated by dividing the signal of the respective VH/VL pair through the signal of the reference IgG1 antibody MOR03080.
CDR-L3 library synthesis by TRIM technology
Trinucleotide (Metkinen Chemistry) containing oligonucleotides were synthesized as previously describedCitation21,Citation28 in collaboration with ELLA Biotech. For synthesis of CDR-L3 cassettes, a total of 15 trimer-phosphoamidite-oligonucleotides with 24 unique, customized TRIMer mixtures were used to create diversity with an amino acid distribution close to that found in natural human rearranged antibodies. CDR-L3 cassettes were assembled and prepared with the appropriate restriction enzymes basically as described before.Citation20,Citation21
CDR-H3 library synthesis by Slonomics technology
The synthesis of the diversified CDR-H3s was performed by using a modified version of the Slonomics technology (MorphoSys) for de novo gene synthesis.Citation29 The adapted procedure allowed the parallel incorporation of complex Anchor molecule mixtures and individual Anchor molecules during elongation reactions, permitting both the generation of variable and constant positions. For the synthesis of the CDR-H3s, a total of 18 different DNA fragments including length variations (from 6 to 17 variable positions) were generated. Using proprietary software, a list of Anchor mixtures with defined ratios and single Anchor molecules according to the amino acid distribution specifications of the CDR-H3 design was created and transferred to a robotic platform for automated pipetting. The diverse DNA sequence fragments were synthesized by using these defined Anchor mixtures in repeated cycles of ligations to precursor molecules followed by subsequent restriction reactions. Assembly of variable sub-fragments was performed by a type IIS restriction endonuclease-based restriction digest and ligation procedure. Upon completion of the assembly, QC was performed for each of the 18 different DNA fragments by separately cloning pools of each fragment and analyzing at least 80 clones by Sanger sequencing each.
Bacterial strains
Electrocompetent E. coli TOP10F′ (Invitrogen) were used for evaluation of display rates. Electrocompetent E. coli MC1061F′ cells with a transformation efficiency of at least 1E+10/µg DNA were used for library cloning. For all other molecular cloning procedures, E.coli XL1-Blue cells (Agilent Technologies) were used and during antibody selections phage were propagated in E.coli TG1F+ cells (Agilent Technologies). Fabs were expressed in E. coli TG1F− cells.
Ylanthia vectors
The pMORPH30 and pMORPHx9 vectorsCitation20 were used as templates for the generation of the new Fab CysDisplay pYPdis10 and Fab expression vectors pJPx1 and pYBex10, respectively, with the following modifications. To increase Fab display levels of the phagemid pYPdis10, codon usage optimization for gIII was performed by GeneArtCitation27 and a D23V mutation was introduced in gIIIp. For both vectors, the ompA and phoA leader sequences upstream of the Ig light and Ig heavy chain encoding sequences were modified at their C-termini to introduce the restriction sites NdeI and NheI, respectively, to assure authentic N-termini of the VL and VH protein sequences and to allow convenient sub-cloning of Fab fragments into pYMex10 or pYMex20 IgG vectors.
Library cloning strategy
Prior to library cloning, the CDR-H3 and CDR-L3 areas of the 12 and 15 different VH and VL region sequences that comprise the finally selected 36 VH/VL pairs were replaced by an 823 bp and 488 bp stuffer fragment containing the β-lactamase and alkaline phosphatase gene, respectively. Stuffer sequences were cloned via BssHII and XhoI (CDR-H3) or BbsI and Acc65I (CDR-L3) into pYPdis10 to facilitate the vector preparations for library cloning.
At first, the diversified CDR-L3 libraries were inserted. Distinct TRIM CDR-L3 library cassettes were prepared for each of the Ig light chain groups Vκ1–05/Vκ1–12, Vκ1–06/Vκ1–09, Vκ1–39, Vκ1–27, Vκ3 and Vλ1–47/Vλ1–51, Vλ2–11/Vλ2–23, Vλ1–40/Vλ3–1 and cloned via BbsI and KpnI into the corresponding pYPdis10 plasmids. After transformation into E. coli MC1061F’ cells and outgrowth for 1 h at 37°C in TB medium, the number of transformed cells was determined by titration on LB/Cam/Gluc agar plates. Amplification of the transformed clones was performed overnight in 20-fold outgrowth volume in LB/Cam/1% Gluc medium at 22°C shaking until an OD600 of 1.8–2.0 was reached and aliquots were stored at −80°C. In addition, DNA was prepared (Qiagen) and the resulting pYPdis10 plasmids containing diversified CDR-L3s were the starting vectors for the diversification of the CDR-H3.
CDR-H3 library cassettes of different lengths (6 to 17 aa) were produced by Slonomics technologyCitation29 as described above and prepared for cloning via restriction sites for BssHII and XhoI. Ligations were used for transformation into E.coli MC1061F’ cells as described above.
Library redundancy analysis and QC by 454 NGS
Quality control of the cloned libraries was performed by high throughput 454 sequencing using the GS Junior System (Roche). In general, amplicon 454 sequencing was performed according to the manuals provided by the manufacturer (GS Junior Titanium Series). Briefly, 4 amplicons of combined Ylanthia sub-libraries were produced by PCR [1 min 98°C, 15 cycles of 98°C for 15 sec, 60°C for 15 sec and 72°C for 15 sec, 72°C for 5 min, 15 ng template DNA, 0.4 μM of each primer, 200 μM dNTPs and 1 U Phusion polymerase (NEB)] using specific amplicon fusion primers. These ~50 nt oligonucleotides enclosed the adaptor A and B sequences, unique sequence tags (multiplex identifiers), as well as target-specific sequences. Agarose gel purified PCR products were further purified using AMPure beads (Beckman Coulter) and quantified by fluorometry using Quant-IT™ PicoGreen® reagent (Invitrogen). Amplicons were pooled, diluted and subjected to an emulsion PCR (emPCR) using the GS Junior Titanium Lib-A Kit (1 copy per bead). Beads were recovered and washed, and DNA positive beads were enriched. Roughly 500,000 enriched beads were deposited on a Pico Titer Plate and the sequencing run was performed as described in the manual.
Results from 454 sequencing were processed using the default amplicon signal processing feature of the GS Junior software package v2.5p1 to obtain high quality, filter passed, sequences. The quality filtered sequences (.fna files) were further analyzed using software developed in-house tailored for processing and analysis of large sequencing data sets.
Large scale phage preparations
Phage preparations were produced as described beforeCitation54 using Hyperphage (Progen). Phage titers were determined by absorbance measurement at OD268 nm (Nanodrop, peqlab) and confirmed by limiting dilution plating of infected E. coli TG1F+ cells on LB/Cam/Gluc plates.
Selection of Fab phage derived from Ylanthia
Equal amounts of phage (~2E+12) from Ylanthia sub-libraries were used for specific antibody selections (“pannings”). Phage were subjected to three rounds of a “solid phase panning” with rhTNFalpha directly immobilized on MaxiSorp™ plates (Nunc) and a “Fc-capture panning” with human IgG1 captured via a Fc fragment specific goat anti-human IgG antibody. Furthermore, “solution pannings” with glyco-biotinylated M-CSF bound to streptavidin coated Dynabeads® (Dynal) and with eGFP, rhErbB4 and rhFZD-4 coupled to carboxy beads (Dynal) were performed according to standard protocols.Citation20,Citation21,Citation54,Citation55 The Fab encoding inserts of the selected pYPdis10 phagemids were subcloned into the bacterial expression vector pYBex_Fab_FH to facilitate expression of soluble Fab in E. coli TG1F− cells and purification via a His6 tag.Citation55
For screening purposes the recombinant antigens were coated onto MaxiSorp™ plates (Nunc) or Neutravidin microtiter plates and incubated with bacterial lysates containing Fab fragments. For detection of antigen-specific Fabs alkaline phosphatase (AP)-conjugated anti-human F(ab’) 2 (Dianova) was applied in combination with AttoPhos (Roche). Fluorescence was measured at an excitation of 440 ± 25 nm and emission of 550 ± 35 nm.
Expression and purification of His6-tagged Fab fragments in E. coli
Expression of unique binding Fab fragments in E. coli TG1F− cells was performed in 500 ml 2× YT cultures containing 34 μg/L Chloramphenicol and 0.1% (w/v) glucose shaken at 22°C until the OD600 nm reached 0.5. Fab expression was induced by the addition of IPTG to a final concentration of 0.5 mM and further cultivation for 20 h at 22°C. Cells were harvested and disrupted using a freeze-thaw cycle followed by lysozyme treatment. His6-tagged Fab fragments were isolated via Profinity IMAC-Resin (Bio-Rad) and eluted using 250 mM imidazole.
Buffer exchange to 1× PBS (pH 7.2, Invitrogen) was performed using PD10 columns (GE Healthcare). Samples were sterile filtered (0.2 μm pore size), and protein concentrations were determined by UV-spectrophotometry (Nanodrop, peqlab). The purity of the samples was analyzed in denaturing, reducing or non-reducing 15% SDS-PAGE (Bio-Rad).
Conversion of Fab fragments into human IgG1 format
To obtain full-length human IgG1 molecules, Fab fragments were subcloned in a two-step cloning procedure from the bacterial Fab expression vector pYBex10 into the mammalian IgG1 expression plasmid pYMex10_h_IgG1, which is a derivative of the pcDNA3.1 and pMORPH4_h_IgG1 vectors.Citation20 In the pYMex10_h_IgG1 vector, the Ig light and Ig heavy chains are encoded on one plasmid.
Expression and purification of full-length human IgG1
Eukaryotic HKB11 cellsCitation77 were transiently transfected with the expression vector pYMex10_h_IgG1. Cell culture supernatants were harvested at day 3 after transfection and IgG1 was purified via Protein A affinity chromatography (MabSelect SURE™, GE Healthcare). Buffer exchange was performed to PBS (pH 7.2, Invitrogen). IgG1 purity was analyzed under denaturing, reducing and non-reducing conditions in SDS-PAGE or by using LabChip® GX II (Caliper/Perkin Elmer). Purified IgG1 concentrations were determined by UV-spectrophotometry (Nanodrop, peqlab). Expression yields were calculated as amount of Protein A purified IgG1 per liter culture supernatant.
Purified Fab and IgG1 thermal stability determination
The thermal stability of purified Fab fragments and IgG1 antibodies was determined by differential scanning fluorometry (DSF).Citation50-Citation52 Diluted Sypro® Orange (Sigma) was added to each well of a 96-well iCycler iQ™ PCR Plate (Bio-Rad), and the Fab or IgG1 samples were tested at a final concentration of at least 0.1 mg/ml on an iQ5™ Thermal cycler (Bio-Rad). The temperature was scanned from 20–95°C at a heating rate of 60°C/h, and the melting temperature of unfolding was calculated by analysis of the inflection point of the fluorescence transition.
Purified Fab and IgG1 monomer content determination
The monomer contents of purified Fab fragments and IgG1 antibodies were determined by size exclusion chromatography (SEC). For separation of Fab fragments, a Superdex™ 75 HR 10/30 column was used on an UltiMate® 3000 Titanium HPLC system (Dionex, Thermo Scientific). For each Fab sample, 10 μg of protein was injected onto the column, separation was performed at a flow rate of 0.05 ml/min with PBS, pH 7.4 (Invitrogen) as running buffer and recorded analyzing the UV absorption at 260 and 280 nm. For separation of IgG1 antibodies, a TSK-Gel® G3000SWxl column (Tosoh Bioscience) was used on an UltiMate® 3000 Titanium HPLC system (Dionex, Thermo Scientific) in combination with miniDAWN™ Treos® and Optilab® rEX (Wyatt Technology) for absolute molecular weight determination. For each IgG1 sample, 15 μg of protein was injected onto the column; separation was performed at a flow rate of 0.5 ml/min and recorded analyzing the UV absorption at 280 nm.
Purified IgG1 low pH stress testing
The ability of the different germline VH/VL pairs in IgG1 format to withstand low pH conditions was tested by incubating the respective samples at a concentration of 2 mg/ml with 1 M citrate buffer at pH 2.3 for 2.5 h and subsequent neutralization using 1 M Tris pH 9.0. Acid stressed and unstressed protein samples were characterized by DSF to evaluate the thermal stability, DLS to assess the hydrodynamic radius and polydispersity in solution, and UV/Vis measurements to determine the concentration of the proteins (280 nm) and the turbidity (320 nm).
Statistics
The distributions of the data were tested for normality using the D’Agostino and Pearson omnibus test. For ease of interpretation, results of non-normally distributed variables are reported as non-transformed values and were analyzed by non-parametric two-tailed Mann–Whitney U-test. Gaussian-distributed parameters were analyzed by two-tailed Student t-test. Multi-group comparison for normally distributed data was performed by the analysis of variance with the Bonferroni post test. All tests were performed using GraphPad Prism® 5.02 software.
| Abbreviations: | ||
| mAbs | = | monoclonal antibodies |
| CDR | = | complementarity determining region |
| Fab | = | fragment antigen binding |
| PTM | = | posttranslational modification site |
| TRIM | = | trinucleotide-directed mutagenesis |
| Ig | = | immunoglobulin |
| HuCAL | = | human combinatorial antibody library |
| VH | = | variable Ig heavy chain region |
| VL | = | variable Ig light chain region |
Additional material
Download Zip (182 KB)Acknowledgments
The authors gratefully acknowledge Tschimegma Bataagijn, Bernadette Gut, Angelika Geide, Christiane Huth, Jan Jaehrling, Tobias Neuber, Andrea Nudlbichler, Solveig Peters, Ingrid Pradel, Olaf Ratsch, and our colleagues from the Protein Sciences Department, as well as from the Slonomics Group for their excellent technical assistance and support. We thank Josef Prassler, Kathrin Tissot, Daniel Weinfurtner, Stefan Pabst and Jan Van den Brulle for highly appreciated input and discussions. Sincere thanks are also given to Kathrin Ladetzki-Baehs and Mark Winderlich for assistance with the preparation of figures and statistics. We thank Josef Prassler, Kathrin Tissot, Margit Urban and Paul Wiegel for critically reading the manuscript.
Disclosure of Potential Conflicts of Interest
All authors were employees of MorphoSys AG during the time of library construction and data evaluation. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed. In connection with this work T.T., T.H., M.E., J.P. and S.U. have a pending patent application, patent number WO2010136598, and T.T., I.S., Y.S. and S.U. have a pending patent application, patent number WO2012066129 and US20120129717.
HuCAL®, HuCAL GOLD®, HuCAL PLATINUM®, CysDisplay® and Ylanthia® are registered trademarks of MorphoSys AG. Slonomics® is a registered trademark of Sloning BioTechnology GmbH, a subsidiary of MorphoSys AG.
Supplemental Material
Supplemental materials may be found here:
http://www.landesbioscience.com/journals/mabs/article/24218
References
- Carter PJ. Potent antibody therapeutics by design. Nat Rev Immunol 2006; 6:343 - 57; http://dx.doi.org/10.1038/nri1837; PMID: 16622479
- Traggiai E, Becker S, Subbarao K, Kolesnikova L, Uematsu Y, Gismondo MR, et al. An efficient method to make human monoclonal antibodies from memory B cells: potent neutralization of SARS coronavirus. Nat Med 2004; 10:871 - 5; http://dx.doi.org/10.1038/nm1080; PMID: 15247913
- Olsson L, Kaplan HS. Human-human hybridomas producing monoclonal antibodies of predefined antigenic specificity. Proc Natl Acad Sci U S A 1980; 77:5429 - 31; http://dx.doi.org/10.1073/pnas.77.9.5429; PMID: 6159646
- Meijer PJ, Andersen PS, Haahr Hansen M, Steinaa L, Jensen A, Lantto J, et al. Isolation of human antibody repertoires with preservation of the natural heavy and light chain pairing. J Mol Biol 2006; 358:764 - 72; http://dx.doi.org/10.1016/j.jmb.2006.02.040; PMID: 16563430
- Tiller T, Meffre E, Yurasov S, Tsuiji M, Nussenzweig MC, Wardemann H. Efficient generation of monoclonal antibodies from single human B cells by single cell RT-PCR and expression vector cloning. J Immunol Methods 2008; 329:112 - 24; http://dx.doi.org/10.1016/j.jim.2007.09.017; PMID: 17996249
- Tiller T. Single B cell antibody technologies. N Biotechnol 2011; 28:453 - 7; http://dx.doi.org/10.1016/j.nbt.2011.03.014; PMID: 21473940
- Köhler G, Milstein C. Continuous cultures of fused cells secreting antibody of predefined specificity. Nature 1975; 256:495 - 7; http://dx.doi.org/10.1038/256495a0; PMID: 1172191
- Lonberg N. Human monoclonal antibodies from transgenic mice. Handb Exp Pharmacol 2008; 69 - 97; http://dx.doi.org/10.1007/978-3-540-73259-4_4; PMID: 18071942
- Hanes J, Plückthun A. In vitro selection and evolution of functional proteins by using ribosome display. Proc Natl Acad Sci U S A 1997; 94:4937 - 42; http://dx.doi.org/10.1073/pnas.94.10.4937; PMID: 9144168
- He M, Taussig MJ. Antibody-ribosome-mRNA (ARM) complexes as efficient selection particles for in vitro display and evolution of antibody combining sites. Nucleic Acids Res 1997; 25:5132 - 4; http://dx.doi.org/10.1093/nar/25.24.5132; PMID: 9396828
- Boder ET, Wittrup KD. Yeast surface display for screening combinatorial polypeptide libraries. Nat Biotechnol 1997; 15:553 - 7; http://dx.doi.org/10.1038/nbt0697-553; PMID: 9181578
- McCafferty J, Griffiths AD, Winter G, Chiswell DJ. Phage antibodies: filamentous phage displaying antibody variable domains. Nature 1990; 348:552 - 4; http://dx.doi.org/10.1038/348552a0; PMID: 2247164
- Smith GP. Filamentous fusion phage: novel expression vectors that display cloned antigens on the virion surface. Science 1985; 228:1315 - 7; http://dx.doi.org/10.1126/science.4001944; PMID: 4001944
- Winter G, Griffiths AD, Hawkins RE, Hoogenboom HR. Making antibodies by phage display technology. Annu Rev Immunol 1994; 12:433 - 55; http://dx.doi.org/10.1146/annurev.iy.12.040194.002245; PMID: 8011287
- Marks JD, Hoogenboom HR, Bonnert TP, McCafferty J, Griffiths AD, Winter G. By-passing immunization. Human antibodies from V-gene libraries displayed on phage. J Mol Biol 1991; 222:581 - 97; http://dx.doi.org/10.1016/0022-2836(91)90498-U; PMID: 1748994
- Vaughan TJ, Williams AJ, Pritchard K, Osbourn JK, Pope AR, Earnshaw JC, et al. Human antibodies with sub-nanomolar affinities isolated from a large non-immunized phage display library. Nat Biotechnol 1996; 14:309 - 14; http://dx.doi.org/10.1038/nbt0396-309; PMID: 9630891
- Huse WD, Sastry L, Iverson SA, Kang AS, Alting-Mees M, Burton DR, et al. Generation of a large combinatorial library of the immunoglobulin repertoire in phage lambda. Science 1989; 246:1275 - 81; http://dx.doi.org/10.1126/science.2531466; PMID: 2531466
- Hoet RM, Cohen EH, Kent RB, Rookey K, Schoonbroodt S, Hogan S, et al. Generation of high-affinity human antibodies by combining donor-derived and synthetic complementarity-determining-region diversity. Nat Biotechnol 2005; 23:344 - 8; http://dx.doi.org/10.1038/nbt1067; PMID: 15723048
- Knappik A, Ge L, Honegger A, Pack P, Fischer M, Wellnhofer G, et al. Fully synthetic human combinatorial antibody libraries (HuCAL) based on modular consensus frameworks and CDRs randomized with trinucleotides. J Mol Biol 2000; 296:57 - 86; http://dx.doi.org/10.1006/jmbi.1999.3444; PMID: 10656818
- Prassler J, Thiel S, Pracht C, Polzer A, Peters S, Bauer M, et al. HuCAL PLATINUM, a synthetic Fab library optimized for sequence diversity and superior performance in mammalian expression systems. J Mol Biol 2011; 413:261 - 78; http://dx.doi.org/10.1016/j.jmb.2011.08.012; PMID: 21856311
- Rothe C, Urlinger S, Löhning C, Prassler J, Stark Y, Jäger U, et al. The human combinatorial antibody library HuCAL GOLD combines diversification of all six CDRs according to the natural immune system with a novel display method for efficient selection of high-affinity antibodies. J Mol Biol 2008; 376:1182 - 200; http://dx.doi.org/10.1016/j.jmb.2007.12.018; PMID: 18191144
- Ponsel D, Neugebauer J, Ladetzki-Baehs K, Tissot K. High affinity, developability and functional size: the holy grail of combinatorial antibody library generation. Molecules 2011; 16:3675 - 700; http://dx.doi.org/10.3390/molecules16053675; PMID: 21540796
- Hoogenboom HR. Selecting and screening recombinant antibody libraries. Nat Biotechnol 2005; 23:1105 - 16; http://dx.doi.org/10.1038/nbt1126; PMID: 16151404
- Hudson PJ. Recombinant antibody fragments. Curr Opin Biotechnol 1998; 9:395 - 402; http://dx.doi.org/10.1016/S0958-1669(98)80014-1; PMID: 9720265
- Söderlind E, Strandberg L, Jirholt P, Kobayashi N, Alexeiva V, Aberg AM, et al. Recombining germline-derived CDR sequences for creating diverse single-framework antibody libraries. Nat Biotechnol 2000; 18:852 - 6; http://dx.doi.org/10.1038/78458; PMID: 10932154
- Jenkins N, Murphy L, Tyther R. Post-translational modifications of recombinant proteins: significance for biopharmaceuticals. Mol Biotechnol 2008; 39:113 - 8; http://dx.doi.org/10.1007/s12033-008-9049-4; PMID: 18327554
- Raab D, Graf M, Notka F, Schödl T, Wagner R. The GeneOptimizer Algorithm: using a sliding window approach to cope with the vast sequence space in multiparameter DNA sequence optimization. Syst Synth Biol 2010; 4:215 - 25; http://dx.doi.org/10.1007/s11693-010-9062-3; PMID: 21189842
- Virnekäs B, Ge L, Plückthun A, Schneider KC, Wellnhofer G, Moroney SE. Trinucleotide phosphoramidites: ideal reagents for the synthesis of mixed oligonucleotides for random mutagenesis. Nucleic Acids Res 1994; 22:5600 - 7; http://dx.doi.org/10.1093/nar/22.25.5600; PMID: 7838712
- Van den Brulle J, Fischer M, Langmann T, Horn G, Waldmann T, Arnold S, et al. A novel solid phase technology for high-throughput gene synthesis. Biotechniques 2008; 45:340 - 3; http://dx.doi.org/10.2144/000112953; PMID: 18778261
- Mietzner B, Tsuiji M, Scheid J, Velinzon K, Tiller T, Abraham K, et al. Autoreactive IgG memory antibodies in patients with systemic lupus erythematosus arise from nonreactive and polyreactive precursors. Proc Natl Acad Sci U S A 2008; 105:9727 - 32; http://dx.doi.org/10.1073/pnas.0803644105; PMID: 18621685
- Tiller T, Tsuiji M, Yurasov S, Velinzon K, Nussenzweig MC, Wardemann H. Autoreactivity in human IgG+ memory B cells. Immunity 2007; 26:205 - 13; http://dx.doi.org/10.1016/j.immuni.2007.01.009; PMID: 17306569
- Tsuiji M, Yurasov S, Velinzon K, Thomas S, Nussenzweig MC, Wardemann H. A checkpoint for autoreactivity in human IgM+ memory B cell development. J Exp Med 2006; 203:393 - 400; http://dx.doi.org/10.1084/jem.20052033; PMID: 16446381
- Wardemann H, Yurasov S, Schaefer A, Young JW, Meffre E, Nussenzweig MC. Predominant autoantibody production by early human B cell precursors. Science 2003; 301:1374 - 7; http://dx.doi.org/10.1126/science.1086907; PMID: 12920303
- Yurasov S, Hammersen J, Tiller T, Tsuiji M, Wardemann H. B-cell tolerance checkpoints in healthy humans and patients with systemic lupus erythematosus. Ann N.Y. Acad.Sci 2005; 1062:165 - 74; http://dx.doi.org/10.1196/annals.1358.019
- Yurasov S, Tiller T, Tsuiji M, Velinzon K, Pascual V, Wardemann H, et al. Persistent expression of autoantibodies in SLE patients in remission. J Exp Med 2006; 203:2255 - 61; http://dx.doi.org/10.1084/jem.20061446; PMID: 16966430
- Brezinschek HP, Foster SJ, Brezinschek RI, Dörner T, Domiati-Saad R, Lipsky PE. Analysis of the human VH gene repertoire. Differential effects of selection and somatic hypermutation on human peripheral CD5(+)/IgM+ and CD5(-)/IgM+ B cells. J Clin Invest 1997; 99:2488 - 501; http://dx.doi.org/10.1172/JCI119433; PMID: 9153293
- Demaison C, David D, Letourneur F, Thèze J, Saragosti S, Zouali M. Analysis of human VH gene repertoire expression in peripheral CD19+ B cells. Immunogenetics 1995; 42:342 - 52; http://dx.doi.org/10.1007/BF00179395; PMID: 7590967
- Foster SJ, Brezinschek HP, Brezinschek RI, Lipsky PE. Molecular mechanisms and selective influences that shape the kappa gene repertoire of IgM+ B cells. J Clin Invest 1997; 99:1614 - 27; http://dx.doi.org/10.1172/JCI119324; PMID: 9120005
- de Wildt RM, Hoet RM, van Venrooij WJ, Tomlinson IM, Winter G. Analysis of heavy and light chain pairings indicates that receptor editing shapes the human antibody repertoire. J Mol Biol 1999; 285:895 - 901; http://dx.doi.org/10.1006/jmbi.1998.2396; PMID: 9887257
- Chothia C, Lesk AM, Gherardi E, Tomlinson IM, Walter G, Marks JD, et al. Structural repertoire of the human VH segments. J Mol Biol 1992; 227:799 - 817; http://dx.doi.org/10.1016/0022-2836(92)90224-8; PMID: 1404389
- Lloyd C, Lowe D, Edwards B, Welsh F, Dilks T, Hardman C, et al. Modelling the human immune response: performance of a 1011 human antibody repertoire against a broad panel of therapeutically relevant antigens. Protein Eng Des Sel 2009; 22:159 - 68; http://dx.doi.org/10.1093/protein/gzn058; PMID: 18974080
- Bhat NM, Bieber MM, Stevenson FK, Teng NN. Rapid cytotoxicity of human B lymphocytes induced by VH4-34 (VH4.21) gene-encoded monoclonal antibodies. Clin Exp Immunol 1996; 105:183 - 90; http://dx.doi.org/10.1046/j.1365-2249.1996.d01-733.x; PMID: 8697629
- Glanville J, Zhai W, Berka J, Telman D, Huerta G, Mehta GR, et al. Precise determination of the diversity of a combinatorial antibody library gives insight into the human immunoglobulin repertoire. Proc Natl Acad Sci U S A 2009; 106:20216 - 21; http://dx.doi.org/10.1073/pnas.0909775106; PMID: 19875695
- Tomlinson IM, Cox JP, Gherardi E, Lesk AM, Chothia C. The structural repertoire of the human V kappa domain. EMBO J 1995; 14:4628 - 38; PMID: 7556106
- Williams SC, Frippiat JP, Tomlinson IM, Ignatovich O, Lefranc MP, Winter G. Sequence and evolution of the human germline V lambda repertoire. J Mol Biol 1996; 264:220 - 32; http://dx.doi.org/10.1006/jmbi.1996.0636; PMID: 8951372
- Dunn-Walters D, Boursier L, Spencer J. Effect of somatic hypermutation on potential N-glycosylation sites in human immunoglobulin heavy chain variable regions. Mol Immunol 2000; 37:107 - 13; http://dx.doi.org/10.1016/S0161-5890(00)00038-9; PMID: 10865109
- Carter P, Presta L, Gorman CM, Ridgway JB, Henner D, Wong WL, et al. Humanization of an anti-p185HER2 antibody for human cancer therapy. Proc Natl Acad Sci U S A 1992; 89:4285 - 9; http://dx.doi.org/10.1073/pnas.89.10.4285; PMID: 1350088
- Ewert S, Huber T, Honegger A, Plückthun A. Biophysical properties of human antibody variable domains. J Mol Biol 2003; 325:531 - 53; http://dx.doi.org/10.1016/S0022-2836(02)01237-8; PMID: 12498801
- Steidl S, Ratsch O, Brocks B, Dürr M, Thomassen-Wolf E. In vitro affinity maturation of human GM-CSF antibodies by targeted CDR-diversification. Mol Immunol 2008; 46:135 - 44; http://dx.doi.org/10.1016/j.molimm.2008.07.013; PMID: 18722015
- Cummings MD, Farnum MA, Nelen MI. Universal screening methods and applications of ThermoFluor. J Biomol Screen 2006; 11:854 - 63; http://dx.doi.org/10.1177/1087057106292746; PMID: 16943390
- Ericsson UB, Hallberg BM, Detitta GT, Dekker N, Nordlund P. Thermofluor-based high-throughput stability optimization of proteins for structural studies. Anal Biochem 2006; 357:289 - 98; http://dx.doi.org/10.1016/j.ab.2006.07.027; PMID: 16962548
- He F, Hogan S, Latypov RF, Narhi LO, Razinkov VI. High throughput thermostability screening of monoclonal antibody formulations. J Pharm Sci 2010; 99:1707 - 20; PMID: 19780136
- Zemlin M, Klinger M, Link J, Zemlin C, Bauer K, Engler JA, et al. Expressed murine and human CDR-H3 intervals of equal length exhibit distinct repertoires that differ in their amino acid composition and predicted range of structures. J Mol Biol 2003; 334:733 - 49; http://dx.doi.org/10.1016/j.jmb.2003.10.007; PMID: 14636599
- Krebs B, Rauchenberger R, Reiffert S, Rothe C, Tesar M, Thomassen E, et al. High-throughput generation and engineering of recombinant human antibodies. J Immunol Methods 2001; 254:67 - 84; http://dx.doi.org/10.1016/S0022-1759(01)00398-2; PMID: 11406154
- Rauchenberger R, Borges E, Thomassen-Wolf E, Rom E, Adar R, Yaniv Y, et al. Human combinatorial Fab library yielding specific and functional antibodies against the human fibroblast growth factor receptor 3. J Biol Chem 2003; 278:38194 - 205; http://dx.doi.org/10.1074/jbc.M303164200; PMID: 12842902
- McGee P. Building better antibody therapeutics. Drug Discovery & Development 2007; 10:26 - 8
- Jayaram N, Bhowmick P, Martin AC. Germline VH/VL pairing in antibodies. Protein Eng Des Sel 2012; 25:523 - 9; http://dx.doi.org/10.1093/protein/gzs043; PMID: 22802295
- Schaefer JV, Plückthun A. Transfer of engineered biophysical properties between different antibody formats and expression systems. Protein Eng Des Sel 2012; 25:485 - 506; http://dx.doi.org/10.1093/protein/gzs039; PMID: 22763265
- Ejima D, Tsumoto K, Fukada H, Yumioka R, Nagase K, Arakawa T, et al. Effects of acid exposure on the conformation, stability, and aggregation of monoclonal antibodies. Proteins 2007; 66:954 - 62; http://dx.doi.org/10.1002/prot.21243; PMID: 17154421
- Low D, O’Leary R, Pujar NS. Future of antibody purification. J Chromatogr B Analyt Technol Biomed Life Sci 2007; 848:48 - 63; http://dx.doi.org/10.1016/j.jchromb.2006.10.033; PMID: 17134947
- Ahrer K, Buchacher A, Iberer G, Jungbauer A. Thermodynamic stability and formation of aggregates of human immunoglobulin G characterised by differential scanning calorimetry and dynamic light scattering. J Biochem Biophys Methods 2006; 66:73 - 86; http://dx.doi.org/10.1016/j.jbbm.2005.12.003; PMID: 16458360
- Brezinschek HP, Foster SJ, Dörner T, Brezinschek RI, Lipsky PE. Pairing of variable heavy and variable kappa chains in individual naive and memory B cells. J Immunol 1998; 160:4762 - 7; PMID: 9590222
- Luo J, Obmolova G, Huang A, Strake B, Teplyakov A, Malia T, et al. Coevolution of antibody stability and Vκ CDR-L3 canonical structure. J Mol Biol 2010; 402:708 - 19; http://dx.doi.org/10.1016/j.jmb.2010.08.009; PMID: 20727359
- Walsh G, Jefferis R. Post-translational modifications in the context of therapeutic proteins. Nat Biotechnol 2006; 24:1241 - 52; http://dx.doi.org/10.1038/nbt1252; PMID: 17033665
- Zheng Y, Tesar DB, Benincosa L, Birnböck H, Boswell CA, Bumbaca D, et al. Minipig as a potential translatable model for monoclonal antibody pharmacokinetics after intravenous and subcutaneous administration. MAbs 2012; 4:243 - 55; http://dx.doi.org/10.4161/mabs.4.2.19387; PMID: 22453096
- Igawa T, Ishii S, Tachibana T, Maeda A, Higuchi Y, Shimaoka S, et al. Antibody recycling by engineered pH-dependent antigen binding improves the duration of antigen neutralization. Nat Biotechnol 2010; 28:1203 - 7; http://dx.doi.org/10.1038/nbt.1691; PMID: 20953198
- Ravn U, Gueneau F, Baerlocher L, Osteras M, Desmurs M, Malinge P, et al. By-passing in vitro screening--next generation sequencing technologies applied to antibody display and in silico candidate selection. Nucleic Acids Res 2010; 38:e193; http://dx.doi.org/10.1093/nar/gkq789; PMID: 20846958
- Venet S, Ravn U, Buatois V, Gueneau F, Calloud S, Kosco-Vilbois M, et al. Transferring the characteristics of naturally occurring and biased antibody repertoires to human antibody libraries by trapping CDRH3 sequences. PLoS One 2012; 7:e43471; http://dx.doi.org/10.1371/journal.pone.0043471; PMID: 22937053
- Kuroda D, Shirai H, Kobori M, Nakamura H. Systematic classification of CDR-L3 in antibodies: implications of the light chain subtypes and the VL-VH interface. Proteins 2009; 75:139 - 46; http://dx.doi.org/10.1002/prot.22230; PMID: 18798566
- Retter I, Althaus HH, Münch R, Müller W. VBASE2, an integrative V gene database. Nucleic Acids Res 2005; 33:Database issue D671 - 4; http://dx.doi.org/10.1093/nar/gki088; PMID: 15608286
- Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 1997; 25:3389 - 402; http://dx.doi.org/10.1093/nar/25.17.3389; PMID: 9254694
- Honegger A, Plückthun A. Yet another numbering scheme for immunoglobulin variable domains: an automatic modeling and analysis tool. J Mol Biol 2001; 309:657 - 70; http://dx.doi.org/10.1006/jmbi.2001.4662; PMID: 11397087
- Hieter PA, Maizel JV Jr., Leder P. Evolution of human immunoglobulin kappa J region genes. J Biol Chem 1982; 257:1516 - 22; PMID: 6276389
- Kawasaki K, Minoshima S, Nakato E, Shibuya K, Shintani A, Schmeits JL, et al. One-megabase sequence analysis of the human immunoglobulin lambda gene locus. Genome Res 1997; 7:250 - 61; http://dx.doi.org/10.1101/gr.7.3.250; PMID: 9074928
- Ravetch JV, Siebenlist U, Korsmeyer S, Waldmann T, Leder P. Structure of the human immunoglobulin mu locus: characterization of embryonic and rearranged J and D genes. Cell 1981; 27:583 - 91; http://dx.doi.org/10.1016/0092-8674(81)90400-1; PMID: 6101209
- Stubenrauch K, Wessels U, Lenz H. Evaluation of an immunoassay for human-specific quantitation of therapeutic antibodies in serum samples from non-human primates. J Pharm Biomed Anal 2009; 49:1003 - 8; http://dx.doi.org/10.1016/j.jpba.2009.01.030; PMID: 19250787
- Cho MS, Yee H, Chan S. Establishment of a human somatic hybrid cell line for recombinant protein production. J Biomed Sci 2002; 9:631 - 8; http://dx.doi.org/10.1007/BF02254991; PMID: 12432229