Abstract
More than half the size of most mammalian genomes is composed by repetitive sequences. Short Interspersed Nuclear Element (SINE) retrotransposons constitute one of the main components of the genomic repetitive fraction. The abundance and evolutionary conservation of these sequences support their contribution to maintain the stability and proper function of the genome. Several recent studies have unveiled some of these intriguing tasks, which include, but are not limited to the control of transcriptional regulation and the organization of the chromatin. Here, we will comment on our recent report characterizing the insulator/boundary activity of a novel B1 SINE retrotransposon (B1-X35S) widely present in the mouse genome. A remarkable finding was that B1-X35S-dependent insulation required not only the combinatorial binding of transcription factors dioxin receptor (AhR) and Snai2/Slug, but also a molecular switch between RNA Polymerases (Pol) Pol III and Pol II. Moreover, B1-X35S seemingly forms heterochromatic barriers next to gene promoters that bioinformatic analyses revealed to dramatically change from embryonic stem (ES) to fibroblasts cells. The vast presence of B1-X35S in the mouse genome (over 14,000 instances) opens the exciting possibility of a complex network in which retrotransposon-derived insulators convert biological input signals into transcriptional responses by defining gene expression domains. The importance of such mechanism in different cellular and physiological processes will be discussed.
Nuclear Organization: The Master Role of CTCF
One of the main concerns for a cell in responding to an external stimulus is to satisfactorily alter its own transcriptome.Citation1 The use of several ‘omic’ techniques, such as gene expression microarrays or chromatin immunoprecipitation (ChIP), followed by massive parallel sequencing (e.g., ChIP-Seq), has greatly increased our knowledge about how the transcriptome and the epigenome is re-shaped under different developmental or physiopathological situations.Citation2 However, many questions still remain regarding the molecular mechanisms through which the eukaryotic genome is dynamically organized into active and inactive expression domains. Despite the partial set of evidences available, it is reasonably well established that the CCCTC-binding factor (CTCF) acts as a master regulator of mammalian genomic boundaries.Citation3 CTCF is able to block the interaction between a transcriptional enhancer and its promoter,Citation4 and to function as a chromatin barrier, thus preventing the spread of repressive heterochromatin along the chromosome.Citation5 These two properties of CTCF have been attributed to its ability to induce the formation of DNA loops that will eventually modulate the accessibility of RNA polymerases and of chromatin modification enzymes to gene regulatory regions,Citation3 likely with the collaboration of additional structural partners such as Cohesin.Citation6,Citation7
Additional reports have shown that CTCF can bind to more than 10,000 sites in mammalian genomes,Citation8 which could reveal a role for this protein in the organization of genome-wide expression patterns. Nevertheless, it should be kept in mind that the actual frequency of CTCF binding to chromatin can be underestimated due to the lack of a fully conserved consensus binding site (probably owing to its binding to variant sequences through combination of several of its eleven zinc-finger domains),Citation3 and a scarce overlapping between sites as observed by ChIP-Seq experiments in different cell lines.Citation8 These facts imply the existence of additional mechanisms governing the recognition of specific DNA sequences by CTCF leading to the dynamic activation of expression domains. In support of this postulate, B1-X35S, which does not have the prototype CTCF element, binds parylated CTCF in vivo and such binding is increased by AhR and Snai2/Slug expression. Because increased recruitment of parylated CTCF to the retrotransposon correlates with insulation and with accumulation of heterochromatic marks upstream of target genes, it is tempting to speculate that AhR + Snai2/Slug modulates the formation of a multimeric protein complex on B1-X35S that includes CTCF and PARP1. Further experiments have to be performed to clarify this issue and to confirm if such mechanism can be extrapolated to other B1-X35S-containing genes and to other physiological and pathological cell conditions.
Retrotransposons as Candidates for Genomic Organizers
Repetitive elements are not just inactivating or deleterious genomic players, as they were considered for a long time.Citation9 Rather, these DNA sequences have the potential to modulate nuclear processes at different levels. For example, thousands of endogenous retroviruses (ERV) are in fact mobile carriers of functional p53 binding sites that are able to regulate the expression of near genes.Citation10 Later, a seminal study by Lunyak et al.Citation11 showed that a B2 SINE retrotransposon located near the Growth Hormone (GH) gene acts as a heterochromatin barrier and CTCF-binding insulator responsible for the GH to be activated in a stage-specific manner during mouse development. Moreover, our studies on the insulator activity of a murine B1-X35S retrotransposonCitation12,Citation13 provide a later example of a genome-wide repetitive element potentially involved in establishing gene expression domains by means of its interaction with transcription factors well known for their contribution to tumor development.
It is interesting that repetitive elements can be transcribed and it was suggested that their resulting RNAs may have a nuclear role in genomic organization. Consistently, human Alu RNAs were shown to repress the transcription of target genes in response to heat-shockCitation14 while both human Alu and murine B2 RNAs disrupted RNA polymerase-promoter interactions with a concomitant decrease in gene expression.Citation15 Albeit additional mechanisms of nuclear organization and gene silencing by Alu retrotransposons recently proposed,Citation16,Citation17 many issues are yet to be investigated about the function of these and of SINE elements in nuclear organization through their potential genome boundary activities. Thus, the functional interaction between insulators/boundaries and transcription factors in defining gene expression and chromatin domains represents a hot question to understand the process of genome-wide control of gene expression.
B1-X35S: A Novel Gene Regulatory Retrotransposon
B1-X35S was described by our group as a new evolutionary subfamily of B1 SINE retrotransposons derived, as human Alus, from the 7SL-RNA.Citation12 Searching for the co-presence in mouse gene promoters of binding sites for two distinct transcription factors, we found an element in which the dioxin receptor binding site (XRE) and the Snai1/2 site (E-box) were simultaneously present and separated by a conserved sequence of 35 base pairs (X35S).Citation12 Remarkably, the amount of gene promoters that contained at least one of these X35S elements was found to exceed 1,300, and from such abundance we thought that they could be functional. In fact, X35S is located within a larger retrotransposon of the B1 family and, interestingly, while the XRE element is common to most B1s, the Snai1/2 site appears de novo in B1-X35S. Evolutionary studies confirmed that B1-X35S was, in fact, a novel young subfamily of the B1 retrotransposon.Citation12 Finally, using in silico analyses and experimental approaches, we began to reveal the contribution of B1-X35S to the transcriptional regulation of target genes. AhR and Snai2/Slug specifically bound this element and such binding provoked the transcriptional repression of downstream genes like Dad1, Lpp and Tbc1d1. Importantly, a similar repressive role for B1-X35S could be predicted genome-wide by in silico analysis of the mouse Gene Expression Atlas database.Citation12
B1-X35S: A Transcription Factor-regulated Genomic Boundary
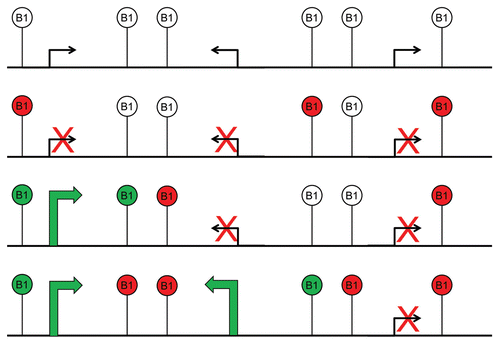
After these compelling results, we sought to analyze whether B1-X35S-dependent repression was due to a silencer/repressive activity or to additional unrelated regulatory processes. We started by considering that gene transcription could be downregulated by two major mechanisms (). On the one hand, B1-X35S could function as a silencer/repressor element triggered by AhR and Snai2/Slug binding to their consensus elements; that possibility supported by the known repressor activities of Snai2/Slug on E-Cadherin during the epithelial-to-mesenchymal transitionCitation18 (EMT) and of AhR on Ltbp1,Citation19 and T-cadherin.Citation20 On the other hand, B1-X35S could function as a boundary/insulator element, whose activity is turned on by AhR and Snai2/Slug binding (). The transcriptional effects derived from an insulator could be confounded with those coming from a repressor and, in fact, several proteins that activate insulators were initially described as transcriptional repressors.Citation3 The discrimination between the silencer vs. the insulator activity of B1-X35S was analyzed in vitro (transfected cells) and in vivo (transgenic zebrafish) using its hypothetical enhancer-blocking function. Strikingly, B1-X35S had a potent intrinsic insulator activity in vitro and in vivo since it could block promoter activity from an enhancer only when located between both regulatory elements (). No less strikingly, overexpression and increased binding of AhR and/or Snai2/Slug improved the insulator activity of B1-X35S in a XRE-specific manner.
As mentioned above, B1-X35S insulation involved basal and AhR + Snai2/Slug-induced binding of parylated CTCF to the element (); if AhR and/or Snai2/Slug have a causal role on such binding is subject of current studies. The insulator activity of the B2-SINE regulating the growth hormone locus during mouse development involved dynamic changes in heterochromatin.Citation11 Suggesting the existence of a common mechanism among different and only partially related SINE retrotransposons, binding of AhR and Snai2/Slug to B1-X35S increased the presence of heterochromatin repressive marks H3K9me3 and H3K27me3 downstream of the element ().
Altogether, these results allowed us to predict the genome-wide role of this retrotransposon in transcriptional insulation. Partial support to that prediction came from in silico screening of public epigenomic datasetsCitation13 containing the required information for the more than 14,000 B1-X35S elements present in the mouse genome. A subset of more than 300 B1-X35S retrotransposons reproduced the H3K9me3 pattern previously determined in specific genes by ChIP assays. Even more interestingly, such pattern, which was evident in embryonic stem (ES), completely disappeared in embryonic fibroblasts. These results open the exciting possibility that B1-X35S, via AhR and Snai2/Slug binding, could modulate chromatin dynamics and nuclear organization of genes relevant for vital cell processes like differentiation. Further studies will be directed to more precisely analyze how chromatin dynamics modulate genome-wide B1-X35S insulation.
Expanding this idea over the genome, it would be important to describe which retrotransposons participate in which cellular processes and how specificities are achieved. The involvement of cell- and developmental stage-specific proteins that bind each insulator element is a potential explanation. A preliminary support for such possibility is provided by Snai1/Snail: this closely related protein that recognizes the same E-box as Snai2/Slug and that binds B1-X35S in vivo, is nevertheless apparently unable to induce its insulator activity.Citation13 AhR and Snail proteins are notable players in tumor induction and progressionCitation18,Citation21 and thus, the implication of B1-X35S in cancer should be studied using different mouse models. Evidences showing that other insulators are relevant for tumor biology,Citation22 and that retrotransposon activity seems to be altered in cancer,Citation23 highlight the importance of this unexplored regulatory pathway.
B1-X35S: The Underlying Molecular Mechanism
Based on the regulatory effects of AhR and Snai2/Slug in B1-X35S-dependent insulation, we decided to analyze the contribution of other proteins to the mechanism. In the case of the B2 SINE retrotransposon, its insulator activity on the growth hormone locus required its own transcription.Citation11 Besides the RNA Pol III promoter that B2 SINE retrotransposons contain, they can also be transcribed (antisense to Pol III) by RNA Pol II.Citation24 These two transcriptional mechanisms participate in regulating the growth hormone gene during mouse development. At early stages, constitutive transcription of the B2 SINE by RNA Pol III helps maintain gene expression turned off. At a specific developmental stage, RNA Pol II binding will increase B2 SINE transcription and promote insulation through chromatin modification and CTCF binding.Citation11 We therefore tested whether B1-X35S-dependent insulation required recruitment of RNA polymerases and if such mechanism was modulated by AhR and Snai2/Slug binding. Surprisingly, we found that AhR and Snai2/Slug binding inversely correlated with Pol III binding,Citation13 that is, those conditions that increase B1-X35S insulation also reduced its transcription by Pol III ().
We speculated that the close proximity between the A and B boxes for RNA Pol III binding and the XRE and E-box elements for AhR and Snai2/Slug binding could produce interference between protein complexes to bind to the retrotransposon. Following that premise, we should expect that AhR and Snai2/Slug binding would decrease B1-X35S transcription due to impaired RNA Pol III recruitment. However, in vitro transcription indicated that increasing AhR binding raised B1-X35S transcription and that this effect was not produced by Snai2/Slug (decreased transcription slightly). A plausible explanation for such apparent discrepancy considers the contribution of other RNA polymerases (e.g., Pol II). Former luciferase experiments could be interpreted assuming that B1-X35S could eventually act as a Pol II promoter.Citation12 The use of RNA Pol III- and Pol II-specific inhibitors readily confirmed that the increase in B1-X35S transcription induced by AhR was Pol II-dependent. Moreover, binding of RNA Pol II and Pol III on B1-X35S is mutually exclusive and, contrary to the B2 SINE of the growth hormone locus, both polymerases transcribe the element in the antisense direction. Interestingly, rather than individually, both RNA polymerases act in concert to trigger B1-X35S insulation. Mutation of the A and B boxes for Pol III binding reduced Pol II-dependent transcription of B1-X35S and impaired its enhancer blocking activity and its response to AhR. We propose a model in which Pol III is initially required at B1-X35S for AhR to induce the recruitment of Pol II (with simultaneous release of Pol III), the increase in transcription and the trigger of insulation.Citation13 Certainly, the common factor for Pol III and Pol II TATA binding protein (TBP), which remains bound to the retrotransposon, could act as a molecular switch for the mechanism. Future studies will address this issue and the implication of other potential intermediate molecules, as the understanding of B1-X35S dynamics will be relevant to analyze genome-wide patterns of gene expression.
Retrotransposons: Insulatory Networks
Putting our work in the current context of the field, it seems plausible that specific types of retrotransposons may play novel roles in organizing and regulating intricate transcriptional pathways in mammals. In such a scenario, different combinations of transcription factors and gene regulatory protein complexes should be able to read the “repetitive elements code” and to provoke transcriptional responses specific for a variety of developmental, environmental, physiological and pathological stimuli (). Integrated, multi-B1 element-dependent insulation might quickly generate very precise transcriptional patterns for the many cellular situations. The notion that rather than useless pieces of DNA, retrotransposons have adapted their functioning to act as genomic traffic lights is rapidly emerging. These dynamic “insulatory networks” may be able to organize gene transcription into expression domains through the collaboration of diverse classes of retrotransposons including SINEs B1 and B2. An additional relevant issue that will be important to analyze is the insulatory potential of retrotransposons contained within the human genome, and their implication in diseases like cancer. In summary, the body of recent reports opens an exciting field in which repetitive elements will be analyzed from a genome-wide perspective in order to decipher their core participation in cell biology.
Abbreviations
| AhR | = | dioxin receptor |
| CTCF | = | CCCTC-binding factor |
| Pol | = | polymerase |
| SINE | = | short-interspersed nuclear element |
Figures and Tables
Figure 1 Silencer/repressor vs. insulator activity of B1-X35S. This scheme shows the functional differences between a silencer/repressor and an insulator/boundary element. A silencer can repress transcription of near genes through repressive marks that spread along the chromatin, and such effect is independent on the relative position of the repressor (upper part). Instead, and insulator blocks the activity of enhancers on cis-promoters but only when located between both regulatory elements (lower part). Represive marks are indicated as filled circles. Vertical black bars on B1-X35S stand for a transcriptional barrier blocking enhancer activity. Crossed arrows indicate lack of transcription while the green arrow represents positive gene expression.
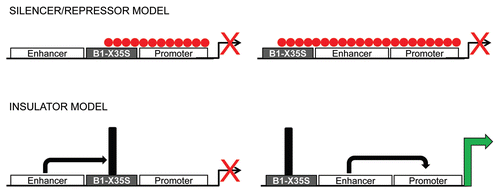
Figure 2 AhR and Snai2/Slug-mediated B1-X35S insulation. The figure summarizes the differences in B1-X35S-dependent insulation under basal and AhR + Snai2/Slug overexpression conditions.
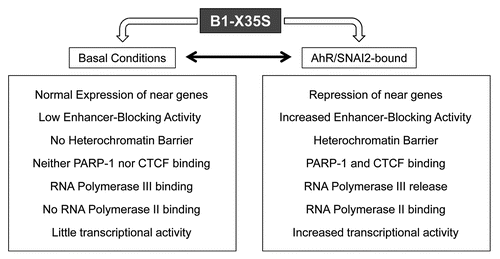
Figure 3 Retrotransposons as Genomic Traffic Lights. The diagram shows some hypothetical transcriptional outputs for three adjacent genes that have several B1 SINE elements located between them. The generation of specific loci models will be needed to study the dynamics of insulatory networks. B1 elements are colored in red or green to indicate inhibition or activation of gene transcription, respectively.
Acknowledgements
This work was supported by grants to P.M.F.S. from the Spanish Ministry of Science and Innovation (MICINN) (SAF2008-00462), the Junta de Extremadura (GRU10008) and the Red Temática de Investigación Cooperativa en Cáncer (RTICC) (RD06/0020/1016, Fondo de Investigaciones Sanitarias (FIS), Carlos III Institute, Spanish Ministry of Health). A.C.R. was an F.P.I. fellow supported by the MICINN. All Spanish funding is co-sponsored by the European Union FEDER program.
References
- Gibson G. The environmental contribution to gene expression profiles. Nat Rev Genet 2008; 9:575 - 581
- Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature 2007; 448:553 - 560
- Phillips JE, Corces VG. CTCF: master weaver of the genome. Cell 2009; 137:1194 - 1211
- Bell AC, West AG, Felsenfeld G. The protein CTCF is required for the enhancer blocking activity of vertebrate insulators. Cell 1999; 98:387 - 396
- Defossez PA, Gilson E. The vertebrate protein CTCF functions as an insulator in Saccharomyces cerevisiae. Nucleic Acids Res 2002; 30:5136 - 5141
- Wendt KS, Yoshida K, Itoh T, Bando M, Koch B, Schirghuber E, et al. Cohesin mediates transcriptional insulation by CCCTC-binding factor. Nature 2008; 451:796 - 801
- Parelho V,, Hadjur S, Spivakov M, Leleu M, Sauer S, Gregson HC, et al. Cohesins functionally associatwith CTCF on mammalian chromosome arms. Cell 2008; 132:422 - 33
- Cuddapah S, Jothi R, Schones DE, Roh TY, Cui K, Zhao K. Global analysis of the insulator binding protein CTCF in chromatin barrier regions reveals demarcation of active and repressive domains. Genome Res 2009; 19:24 - 32
- Goodier JL, Kazazian H Jr. Retrotransposons revisited: the restraint and rehabilitation of parasites. Cell 2008; 135:23 - 35
- Wang T, Zeng J, Lowe CB, Sellers RG, Salama SR, Yang M, et al. Species-specific endogenous retroviruses shape the transcriptional network of the human tumor suppressor protein p53. Proc Natl Acad Sci USA 2007; 104:18613 - 18618
- Lunyak VV, Prefontaine GG, Nunez E, Cramer T, Ju BG, Ohgi KA, et al. Developmentally regulated activation of a SINE B2 repeat as a domain boundary in organogenesis. Science 2007; 317:248 - 251
- Roman AC, Benitez DA, Carvajal-Gonzalez JM, Fernandez-Salguero PM. Genome-wide B1 retrotransposon binds the transcription factors dioxin receptor and Slug and regulates gene expression in vivo. Proc Natl Acad Sci USA 2008; 105:1632 - 1637
- Roman AC, Gonzalez-Rico FJ, Molto E, Hernando H, Neto A, Vicente-Garcia C, et al. Dioxin receptor and SLUG transcription factors regulate the insulator activity of B1 SINE retrotransposons via an RNA polymerase switch. Genome Res 2011; 21:422 - 432
- Mariner PD, Walters RD, Espinoza CA, Drullinger LF, Wagner SD, Kugel JF, et al. Human Alu RNA is a modular transacting repressor of mRNA transcription during heat shock. Mol Cell 2008; 29:499 - 509
- Yakovchuk P, Goodrich JA, Kugel JF. B2 RNA and Alu RNA repress transcription by disrupting contacts between RNA polymerase II and promoter DNA within assembled complexes. Proc Natl Acad Sci USA 2009; 106:5569 - 5574
- Chen LL, DeCerbo JN, Carmichael GG. Alu element-mediated gene silencing. EMBO J 2008; 27:1694 - 1705
- Shen S, Lin L, Cai JJ, Jiang P, Kenkel EJ, Stroik MR, et al. Widespread establishment and regulatory impact of Alu exons in human genes. Proc Natl Acad Sci USA 2011; 108:2837 - 2842
- Peinado H, Olmeda D, Cano A. Snail, Zeb and bHLH factors in tumour progression: an alliance against the epithelial phenotype?. Nat Rev Cancer 2007; 7:415 - 428
- Gomez-Duran A, Ballestar E, Carvajal-Gonzalez JM, Marlowe JL, Puga A, Esteller M, et al. Recruitment of CREB1 and histone deacetylase 2 (HDAC2) to the mouse Ltbp-1 promoter regulates its constitutive expression in a dioxin receptor-dependent manner. J Mol Biol 2008; 380:1 - 16
- Niermann T, Schmutz S, Erne P, Resink T. Aryl hydrocarbon receptor ligands repress T-cadherin expression in vascular smooth muscle cells. Biochem Biophys Res Commun 2003; 300:943 - 949
- Barouki R, Coumoul X, Fernandez-Salguero PM. The aryl hydrocarbon receptor, more than a xenobiotic-interacting protein. FEBS Lett 2007; 581:3608 - 3615
- Witcher M, Emerson BM. Epigenetic silencing of the p16(INK4a) tumor suppressor is associated with loss of CTCF binding and a chromatin boundary. Mol Cell 2009; 34:271 - 284
- Garcia-Perez JL, Morell M, Scheys JO, Kulpa DA, Morell S, Carter CC, et al. Epigenetic silencing of engineered L1 retrotransposition events in human embryonic carcinoma cells. Nature 2010; 466:769 - 773
- Ferrigno O, Virolle T, Djabari Z, Ortonne JP, White RJ, Aberdam D. Transposable B2 SINE elements can provide mobile RNA polymerase II promoters. Nat Genet 2001; 28:77 - 81