Abstract
A large portion of the mammalian genome is derived from ancient transposable elements. Retroelements, transported by an intracellular copy-and-paste process involving an RNA intermediate (retrotransposition), constitute a majority of these mobile genetic elements. Endogenous retroviruses are LTR-type retroelements accounting for around 8% of human or murine genomic DNA. Non-LTR members are present in extremely high copy numbers; with LINE-1 contributing to nearly 40% of human and murine genomes. These LINE-1 elements modify mammalian genomes not only through insertions, but also by indirect replication of nonautonomous retrotransposons such as SINEs. As expected, cellular machineries of vertebrate’s innate immunity have evolved to support a balance between retroelement insertions that cause deleterious gene disruptions and those that confer beneficial genetic diversity. The ability of APOBEC3 cytidine deaminases targeting DNA to restrict a broad number of retroviruses and retroelements is now well established. More recently, the RNA editing family member APOBEC1, a protein involved in lipid transport, has also been shown to be involved in keeping mobile elements under control. This review discusses current understanding of the mechanism of action of the AID/APOBEC family, and their role in controlling the retrotransposition of endogenous retroelements.
Introduction
The ability of the AID/APOBEC family proteins to confer intrinsic immunity to retroviral infection was first recognized in the case of human APOBEC3G (hA3G), which can block the replication of HIV-1 mutant lacking the vif gene.Citation1 Members of the APOBEC3(A3) cytidine deaminases are innate restriction factors that act against two distinct endogenous retroelements: LTR retrotransposons (or endogenous retroviruses) and non-LTR retroelements such as LINE-1 (L1) (reviewed in refs. Citation2 and Citation3). Endogenous retroviruses, which are structurally similar to HIV-1 and other infectious retroviruses, are inherited like cellular genes following germ-line transmission of some retroviral genomes, accumulating throughout evolution in many organisms. Endogenous retroviruses, also called extrachromosomally-primed (EP) retrotransposons (reviewed in refs. Citation4), pre-dominantly undergo reverse transcription in the cytoplasm of infected cells (). On the other hand, the replication cycle of the non-LTR retrotransposon, also called target-primed (TP) retrotransposons, is different, with reverse transcription occuring exclusively within the nucleus. These autonomous EP and TP retrotransposons modify mammalian genomes not only by creating insertions, but also by indirect replication of SINEs and processed pseudogenes.
A3 molecules belong to a family of proteins that also includes APOBEC1 (A1), AID, APOBEC2 (A2) and APOBEC4 (A4). These proteins mediate the enzymatic conversion of cytosine bases to uracils (C to U) on DNA and/or RNA. A1, the catalytic component of a complex that deaminates apo B mRNA in gastrointestinal tissues, is the original member of this family and remains the best characterized. AID, a DNA-editing enzyme that is the second member to be identified, has been shown to act on the immunoglobulin locus to trigger the antigen-dependent diversification processes in activated B cells (reviewed in ref. Citation5). A2 is ubiquitously present in vertebrates and expressed primarily in cardiac and skeletal muscle,Citation6 but the physiological function of A2 remains unknown. A4 is present in mammals, birds, amphibian, but not in fishes.Citation7 In mammals, A4 is expressed most prominently in testis, suggesting its role in spermatogenesis.
In this communication, we briefly summarize advances in the general knowledge of AID/APOBEC family proteins as guardians of the cell genome against invading endogenous retroelements. In particular, we describe the role played by A1 cytidine deaminase in this intrinsic immunity in mammalian species, in addition to its integral role in apo B mRNA editing.
AID/APOBEC Deaminases as Restriction Factors of Endogenous Retroviruses
Mammalian endogenous retroviruses are multicopy retroelements accounting for around 8% of human or murine genomic DNA, with most having been inactivated over time through mutations and deletions (reviewed in ref. Citation8 and Citation9). These ancestral retroelements may have been deleterious, being implicated in having immunomodulatory and tumorgenic potentials to cause several diseases in their host. But it is obvious that the considerable number of endogenous retroviruses inserted in ancestral mammalian chromosomes has had a profound impact on the landscape and plasticity of the host genome, increasing significantly the rate of primate evolution.
Many endogenous retroviruses entered the germ line as infectious retroviruses at several time points in the ancestral line of primate evolution. None of the endogenous retroviruses currently present in the human genome (HERVs), however, is replication competent. Several, including some murine IAP and MusD sequences, are still mobile, capable of retrotransposition in the host genome. Following the discovery of the restriction activities of A3 proteins against HIV-1, similar DNA editing-dependent activity against endogenous retrovirus was reported in references Citation10 and Citation11. The AID/APOBEC family proteins contain either one or two copies of the active site core motif His-X-Glu-X23–28-Pro-Cys-X2–4-Cys, in which the His and two Cys residues coordinate Zn2+ ion, and Glu functions as the catalytic residue. The inhibitory activities against IAP and MusD observed with A3 proteins appeared to be based, at least in part, on cytidine deamination. Consistent with reports that the inhibitory activity against exogenous retrovirus such as HIV-1 by A3 proteins was mediated by their selective incorporation into retroviral particle through interaction with the Gag protein, direct interactions between A3 proteins and IAP Gag had been demonstrated. Further, genetic analyses indicated that the molecular mechanism(s) for the editing of reversetranscribed DNA from endogenous retrovirus and exogenous retrovirus such as HIV-1 overlapped.Citation10,Citation11 This intra-cellular editing mechanism induces degradation of the nascent viral intermediates before integration, possibly due to the base excision repair pathway, a process that is initiated by cellular uracil-DNA glycosylases and can lead to fatal mutagenesis.
Subsequently, the single-domain cytidine deaminase AID from multiple species including primary vertebrates, and A1 proteins from several mammalian species were also found to possess the capacity to inhibit endogenous retroviruses.Citation12–Citation14 In addition, several reports indicated that A1s from several mammalian species inhibit a wide range of exogenous retroviruses such as HIV-1.Citation15–Citation18 These results raise the possibility that not only A3 proteins but also AID and A1 cytidine deaminases are likely to participate in the intrinsic immunity in various mammalian species against the retrotransposition of endogenous and exogenous retroviruses. The catalytic activity of A1s appeared to be critical for these repressive potentials.Citation14,Citation17 In addition, although the expression of A1 in human has only been formally documented in gastrointestinal tissue, the distribution for mouse, rat and rabbit A1s is much wider, including tissues such as hepatocytes and spleen which are assumed to have little apoB mRNA. Furthermore, A1 mRNA is expressed in ovarian and testis tissue, at least in mouse and rabbit, placing A1 in a compartment where endogenous retrovirus may have the greatest impact in vivo.Citation14
Interestingly, the powerful inhibition of endogenous retrovirus by the single-domain cytidine deaminase hA3A appeared to occur independently of editing.Citation11 Nevertheless, sequence analysis of the retrotransposed DNA copies using a differential DNA denaturation PCR (3D-PCR) to detect G-to-A hypermutated proviral genome revealed extensive editing in the presence of hA3A.Citation14 Thus, the inhibitory mechanism(s) used by hA3A to inhibit retrotransposition of endogenous retrovirus is currently not fully understood, and remains controversial.
AID/APOBEC Deaminases as Restriction Factors of Non-LTR Retrotransposons
A large portion of the mammalian genome is composed of non-LTR retrotransposons, with L1 elements contributing to over 35% of the mass of the human and other mammalian genomes. Furthermore, around 100 copies of L1s in humans and 3,000 copies in mice appear to be active (reviewed in refs. Citation8 and Citation9). These elements modify mammalian genomes not only by insertions, but also by indirect replication of SINEs that include the most prominent and active member Alu in human or B1 in mice. L1 retrotransposition has been demonstrated to result in human diseases such as Haemophiliae VIII/IX and Duchene muscular dystrophy, and in the generation of novel polymorphisms.
Members of the A3 cytidine deaminases are certainly one class of the cellular machineries that play roles as innate restriction factors against these non-LTR retrotransposons (reviewed in refs. Citation2 and Citation3). All human A3 family proteins, hA3A-hA3H inhibit autonomous retroelement L1 to varying degree.Citation19,Citation20 hA3A and hA3B are the most potent inhibitors of L1s, although hA3A does not exhibit any activity against HIV-1.
In addition, hA3A, hA3B, hA3C and hA3G were found to inhibit L1-mediated Alu retrotransposition. The mechanism of antiretroviral and antiretrotransposon inhibition differs and the latter appears to be independent of the enzymatic activity. Of note, although A3A inhibits L1 retrotransposition efficiently without any apparent DNA deamination, the conserved catalytic Glu in the single-domain A3A cytidine deaminase appears to be critical for its activity against L1, suggesting the importance for proper folding and target factors such as RNA or protein interaction in addition to the cytidine deamination.Citation20 hA3A has been suggested to affect transgene expression through extensive deamination of transfected plasmid DNA.Citation21 Thus, it is likely that hA3A utilizes a mechanism different from those observed in other A3 proteins for inhibiting the endogenous retroelements.
In contrast to A3 proteins, it has been suggested that neither AID nor A1 possess any activity against the L1 retrotranspositions. However, similar DNA editing-independent anti-L1 activity was recently documented for AID from multiple species.Citation13 Furthermore, A1 proteins from several mammalian species were also found to possess the capacity to inhibit the L1 retrotranspositions.Citation14 The anti-L1 activity of A1 appears to be much more profound than those documented for AID, and also does not require cytidine deaminase activity. The replication cycle of the non-LTR retrotransposon L1s is different from that of LTR retroelements such as IAP or MusD, in which reverse transcription occurs within the cytoplasm (). So far, similar DNA editing-independent L1 restriction machineries have been confirmed for a variety of AID/APOBEC family proteins, but the exact step(s) of L1 replication affected has yet to be determined. Whether these AID/APOBEC proteins bind to L1 encoded ORFs and/or specific sequences in L1 RNA and/or host factor(s) that facilitate retrotransposition remains to be determined. A1 as well as hA3A and hA3G are able to inhibit nascent L1 DNA accumulation, suggesting interference with L1 reverse transcription, integration and/or intracellular movement of L1 ribonucleoprotein particles (RNPs).Citation14,Citation19 Furthermore, the suppressive activity against de novo L1 DNA synthesis was mainly in a deamination-independent manner, and was not affected by subcellular localization of the proteins.Citation14 AID/APOBEC proteins might be able to interact with the cytoplasmic RNPs and interfere with subsequent RNPs transport and/or nuclear import. Alternatively, AID/APOBEC proteins may function at other steps downstream of RNPs formation in the L1 retrotransposition pathway. For example AID/APOBEC proteins in the nucleus might be able to access to L1 RNA and block the generating nick or subsequent priming of reverse transcription of 1st strand, by interfering the access of the L1 RNA to the target DNA. AID/APOBEC proteins without nuclear localizing signal (NLS) might be able to enter the nucleus with the RNP complex in which ORF2p harbors a putative NLS. The interactions between AID/APOBEC proteins and ORF2p have been suggested, however, the direct interaction has not been demonstrated, thus far.
Indeed, an interaction between A1 as well as hA3A and hA3G proteins with L1 and cellular RNAs such as polymerase II-transcribed GAPDH and elF4G2 RNA, polymerase III-derived 7SL RNA had been documented.Citation14 The copy numbers of L1 and cellular RNAs with A1s and hA3A were found to be lower than those seen with hA3G. It is plausible that this weaker interaction can be explained by the fact that A1s and hA3A are single-domain, while hA3G have double deamination domains. hA3G has been documented to interact with cellular RNAs to assemble into high-molecular-mass (HMM) ribonucleoprotein complexes that converts to a low-molecular-mass (LMM) form by RNase treatment, and to associate with stress granules, staufen granules or P bodies (reviewed in ref. Citation3). However, it appeared that the inhibitory activity of human A3 proteins against L1 retrotransposition does not correlate with intracellular HMM formation or P-body association.Citation20 A1 proteins were found to exist in a HMM form both in the absence or presence of L1. Notably, and in sharp contrast to hA3G, A1 distribution was not affected by RNase treatment, suggesting that this single-domain cytidine deaminase may be able to interact differently and/or more strongly with host RNA(s). The exact step(s) of L1 replication affected by these AID/APOBEC proteins remains to be verified, in order to clarify the molecular mechanism through which L1 retrotranspositions are inhibited, mainly by the deamination-independent manner. Elucidation of these deamination-independent repressive activities of AID/APOBEC proteins to L1 retrotranspositions may provide a new insights into the consequences of deamination-independent HIV-1 inhibition by A3 proteins.
Despite the impact of L1 insertion on mammalian genome evolution, much of the process of L1 retrotransposition, especially in vivo remains unexplored. Recent studies suggested that de novo L1 retrotransposition usually occurs early in embryogenesis.Citation22 This scenario indicated that the germ cells should have evolved several post-transcriptional defense mechanisms that strictly prevent L1 integration into the genome. These might include posttranscriptional silencing via RNA interference (RNAi),Citation23 and cytidine deamination via the A3 family-mediated machinery.Citation10,Citation11 In addition to these well documented defense mechanisms, recent studies including ours indicate that the AID- and A1-mediated machineries may contribute to control L1 retrotransposition in germ cells as well.
Evolution and Diversification of AID/APOBEC Family
AID/APOBEC family proteins targeting nucleic acids are thought to arise at the beginning of the vertebrate radiation and further expanded in mammalian species.
The single-domain cytidine deaminase of AID as well as A2 is likely to be the evolutionary precursor, since only AID- and A2-like but not A3-related sequences appear to be encoded in the genomes of non-mammalian primary vertebrates such as birds and fishes (reviewed in ref. Citation24). Thus, A1 and the cluster of A3 gene(s) is likely to have arisen from duplication of the locus for the AID- and A2-like ancestral gene through a series of gene duplication and diversification.
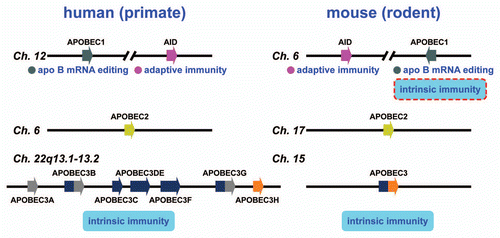
The human genome encodes seven A3 proteins, from A3A to A3H, while the mouse genome contains a single A3 gene (). The A3 genes encode both single- and double-domain cytidine deaminases, and appear to be specific to placental (eutherian) mammals. The ancestral single-domain fusion or duplication is thought to have occurred after the divergence of placental and non-placental mammals. This is supported by the fact that non-placental mammals such as opossum (marsupialia) and platypus (monotremate) genomic sequences appear to lack A3 genes.Citation25 Thus, the double-domain cytidine deaminases have been identified only in placental mammals, thus far. In contrast, the single-domain A1 protein with apoB mRNA editing activity appears to be expressed in the marsupial opossum.Citation26 The recent study further indicated that the genomes of non-mammalian vertebrates such as lizard and zebra finch encode A1-like molecules.Citation27 Interestingly, the identified A1-like enzyme appears to lack a C-terminal region important for mammalian apoB mRNA editing. Indeed, apoB mRNA was not edited by this A1-like molecules in these primary vertebrates, however, this lizard A1-like molecule is able to deaminate DNA in bacteria. These observations raise the possibility that the ability of single-domain cytidine deaminase A1 to edit RNA has been a later evolutionary event well after their divergence from a common ancestor of DNA mutators, and that A1-like molecules encoded by lizard and zebra finch genomes may play roles in restricting the retrotransposition of endogenous retroelements.
The spectrum of biological functions of the AID/APOBEC family (comprising AID, A1, A2, A3 subgroups, A4 and more) of DNA/RNA cytidine deaminases in vertebrates is expanding. Clearly, uncontrolled expression or transposition of autonomous LTR and non-LTR retrotransposons is deleterious for the host by causing lethal gene disruptions. But appropriate levels of retrotransposition in germline cells and/or early in embryogenesis might contribute to beneficial genetic diversity and host genome evolution. It is tempting to speculate that the mammalian A3 genes have been under strong positive selection over hundred million years of mammalian evolution. It is noteworthy that the rapid expansion of the A3 gene family occurred in primates.Citation28,Citation29 This expansion may have caused the dramatic decline of retrotransposon activities in primates. The function(s) of A1 as an intrinsic immunity is taken over by expanded A3s in human, but is conserved in rodent and rabbit.
In summary, several members of the AID/APOBEC family proteins appear to play important roles in the intrinsic immune mechanisms for preventing the spread of foreign as well as endogenous nucleic acids through both editing and non-editing mechanisms. Further investigation into the activities against the endogenous retroelements should provide insights into the evolution and diversification of AID/APOBEC family in mammalian species.
Abbreviations
| APOBEC | = | apolipoprotein B mRNA-editing, catalytic polypeptide |
| apo B | = | apolipoprotein B |
| AID | = | activation-induced cytidine deaminase |
| HIV-1 | = | human immunodeficiency virus type 1 |
| LTR | = | long-terminal-repeat |
| LINE | = | long-interspersed nuclear element |
| SINE | = | short-interspersed nuclear element |
| HERV | = | human endogenous retroviruse |
| IAP | = | intracisternal A particle |
| VLP | = | virus-like particle |
Figures and Tables
Figure 1 Models of the retroelement retrotransposition cycle. (Left part) Retrotransposition pathway of endogenous retroviruses (or LTR retrotransposons) such as murine IAP and MusD. Mammalian endogenous retroviruses sequences are structurally similar to infectious retroviruses such as HIV-1 and MLV. The infectious retroviruses encode an envelope protein (Env) that facilitates their transmission from one cell to another, whereas endogenous retroviruses either lack or contain a remnant of an env gene and can only integrate into the genome at a new site within their cell of origin. Endogenous retroviruses also contain slightly overlapping ORFs for their group-specific antigen (Gag), protease (Prt), polymerase (Pol) and terminal LTRs. The pol genes encode a reverse transcriptase, ribonuclease H and integrase to provide enzymatic activities for generating proviral cDNA from viral genomic RNA and inserting it into the host genome. Their life cycle includes the formation of VLPs, which remain intracellular. Reverse transcription of endogenous retrovirus genomic RNA occurs within the VLP in the cytoplasm, and is a complicated, multistep process. (Right part) Retrotransposition pathway of L1 retroelements. A ∼6 kb functional L1 element contains an internal RNA polymerase II promoter in the 5′ UTR , followed by two open reading frames. ORF1 encodes RNA-binding protein (ORF1p) that is required for ribonucleoprotein particle (RNP) formation in the cytoplasm. ORF2 encodes a protein with endonuclease and reverse transcriptase activities (ORF2p). Both ORF1p and ORF2p are critical for retrotransposition by a “copy and paste” mechanism. A short 3′ UTR is followed by a poly(A) tail, and the entire element is flanked by target site duplications (TSD). L1 DNA synthesis in the nucleus is based on “target-primed reverse transcription (TPRT)” in which ORF2p nicks target chromosomal DNA, using the resultant 3′-OH to prime the reverse transcription of L1 RNA as a template. At an early phase of replication, L1 RNA forms a RN P complex in the cytoplasm as a retrotransposition intermediate by associating with ORF1p and ORF2p.
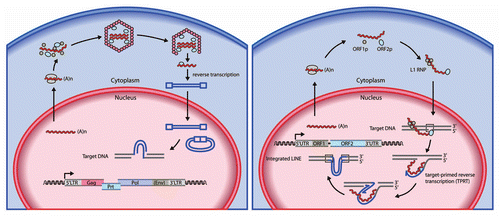
Figure 2 AID/APOBEC genes in human and murine genomes. A schematic of the human and murine genomes containing members of the AID/APOBEC family. There are several forms of mammalian AID/APOBEC proteins with distinct functions in vivo: activation-induced deaminase (AID), APOBEC1(A1), APOBEC2(A2), APOBEC3(A3) and APOBEC4(A4). The primate-specific cluster of at least seven A3 related genes: A3A-3H, resides on the same chromosome within ∼150 kb. In contrast, mice retains only a single A3 gene, located on chromosome 15e2, indicating a relatively recent, and possibly unprecedented gene expansion has occurred in mammalian species. Each A3 gene encodes a protein with one or two conserved zinc-coordinating motifs (Z1, Z2 or Z3).Citation25,Citation30 Grey and blue denote Z1 and Z2, respectively, while orange denotes Z3. Rodents encode only one A3 gene (a Z2–Z3 fusion). A1 and AID are located approximately 900 kb apart on human chromosome 12.
Acknowledgments
We thank C. Cheng-Mayer for reviewing the article. This work was supported by Higo Bank.
References
- Sheehy AM, Gaddis NC, Choi JD, Malim MH. Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral Vif protein. Nature 2002; 418:646 - 650; PMID: 12167863; http://dx.doi.org/10.1038/nature00939
- Holmes RK, Malim MH, Bishop KN. APOBEC-mediated viral restriction: not simply editing?. Trends Biochem Sci 2007; 32:118 - 128; PMID: 17303427; http://dx.doi.org/10.1016/j.tibs.2007.01.004
- Chiu YL, Green WC. The APOBEC3 cytidine deaminases: an innate defensive network opposing exogenous retroviruses and endogenous retro-elements. Annu Rev Immunol 2008; 26:317 - 353; PMID: 18304004; http://dx.doi.org/10.1146/annurev.immunol.26.021607.090350
- Beauregard A, Curcio MJ, Belfort M. The take and give between retrotransposable elements and their hosts. Annu Rev Genet 2008; 42:587 - 617; PMID: 18680436; http://dx.doi.org/10.1146/annurev.genet.42.110807.091549
- Wedekind JE, Dance GSC, Sowden MP, Smith HC. Messenger RNA editing in mammals: new members of the APOBEC family seeking roles in the family business. Trends Genet 2003; 19:207 - 216; PMID: 12683974; http://dx.doi.org/10.1016/S0168-9525(03)00054-4
- Liao W, Hong SH, Chan BH, Rudolph FB, Clark SC, Chan L. APOBEC-2, a cardiac- and skeletal muscle-specific member of the cytidine deaminase supergene family. Biochem Biophys Res Commun 1999; 260:398 - 404; PMID: 10403781; http://dx.doi.org/10.1006/bbrc.1999.0925
- Rogozin IB, Basu MK, Jordan IK, Pavlov YI, Koonin EV. APOBEC4, a new member of the AID/APOBEC family of polynucleotide (deoxy)cytidine deaminases predicted by computational analysis. Cell Cycle 2005; 4:1281 - 1285; PMID: 16082223; http://dx.doi.org/10.4161/cc.4.9.1994
- Bannert N, Kurth R. Retroelements and the human genome: new perspectives on an old relation. Proc Natl Acad Sci USA 2004; 101:14572 - 14579; PMID: 15310846; http://dx.doi.org/10.1073/pnas.0404838101
- Kazazian HH Jr. Mobile elements: drivers of genome evolution. Science 2004; 303:1626 - 1632; PMID: 15016989; http://dx.doi.org/10.1126/science.1089670
- Esnault C, Heidmann O, Delebecque F, Dewannieux M, Ribet D, Hance AJ, et al. APOBEC3G cytidine deaminase inhibits retrotransposition of endogenous retroviruses. Nature 2005; 433:430 - 433; PMID: 15674295; http://dx.doi.org/10.1038/nature03238
- Bogerd HP, Wiegand HL, Doehle BP, Lueders KK, Cullen BR. APOBEC3A and APOBEC3B are potent inhibitors of LTR-retrotransposon function in human cells. Nucleic Acids Res 2006; 34:89 - 95; PMID: 16407327; http://dx.doi.org/10.1093/nar/gkj416
- Esnault C, Millet J, Schwartz O, Heidmann T. Dual inhibitory effects of APOBEC family proteins on retrotransposition of mammalian endogenous retroviruses. Nucleic Acids Res 2006; 34:1522 - 1531; PMID: 16537839; http://dx.doi.org/10.1093/nar/gkl054
- MacDuff DA, Demorest ZL, Harris RS. AID can restrict L1 retrotransposition suggesting a dual role in innate and adaptive immunity. Nucleic Acids Res 2009; 37:1854 - 1867; PMID: 19188259; http://dx.doi.org/10.1093/nar/gkp030
- Ikeda T, Abd El Galil KH, Tokunaga K, Maeda K, Sata T, Sakaguchi N, et al. Intrinsic restriction activity by apolipoprotein B mRNA editing enzyme APOBEC1 against the mobility of autonomous retrotransposons. Nucleic Acids Res 2011; 39:5538 - 5554; PMID: 21398638; http://dx.doi.org/10.1093/nar/gkr124
- Bishop KN, Holmes RK, Sheehy AM, Malim MH. APOBEC-mediated editing of viral RNA. Science 2004; 305:645; PMID: 15286366; http://dx.doi.org/10.1126/science.1100658
- Bishop KN, Holmes RK, Sheehy AM, Davidson NO, Cho SJ, Malim MH. Cytidine deamination of retroviral DNA by diverse APOBEC proteins. Curr Biol 2004; 14:1392 - 1396; PMID: 15296758; http://dx.doi.org/10.1016/j.cub.2004.06.057
- Ikeda T, Ohsugi T, Kimura T, Matsushita S, Maeda Y, Harada S, et al. The antiretroviral potency of APOBEC1 deaminase from small animal species. Nucleic Acids Res 2008; 36:6859 - 6871; PMID: 18971252; http://dx.doi.org/10.1093/nar/gkn802
- Petit V, Guétard D, Renard M, Keriel A, Sitbon M, Wain-Hobson S, et al. Murine APOBEC1 is a powerful mutator of retroviral and cellular RNA in vitro and in vivo. J Mol Biol 2009; 385:65 - 78; PMID: 18983852; http://dx.doi.org/10.1016/j.jmb.2008.10.043
- Kinomoto M, Kanno T, Shimura M, Ishizaka Y, Kojima A, Kurata T, et al. All APOBEC3 family proteins differentially inhibit LINE-1 retrotransposition. Nucleic Acids Res 2007; 35:2955 - 2964; PMID: 17439959; http://dx.doi.org/10.1093/nar/gkm181
- Niewiadomska AM, Tian C, Tan L, Wang T, Sarkis PTN, Yu XF. Differential inhibition of long interspersed element 1 by APOBEC3 does not correlate with high-molecular-mass-complex formation or P-body association. J Virol 2007; 81:9577 - 9583; PMID: 17582006; http://dx.doi.org/10.1128/JVI.02800-06
- Stenglein MD, Burns MB, Li M, Lengyel J, Harris RS. APOBEC3 proteins mediate the clearance of foreign DNA from human cells. Nat Struct Mol Biol 2010; 17:222 - 229; PMID: 20062055; http://dx.doi.org/10.1038/nsmb.1744
- Kano H, Godoy I, Courtney C, Vetter MR, Gerton GL, Ostertag EM, et al. L1 retrotransposition occurs mainly in embryogenesis and creates somatic mosaicism. Genes Dev 2009; 23:1303 - 1312; PMID: 19487571; http://dx.doi.org/10.1101/gad.1803909
- Yang N, Kazazian HH Jr. L1 retrotransposition is suppressed by endogenously encoded small interfering RNAs in human cultured cells. Nat Struct Mol Biol 2006; 13:763 - 771; PMID: 16936727; http://dx.doi.org/10.1038/nsmb1141
- Conticello SG, Thomas CJF, Petersen-Mahrt SK, Neuberger MS. Evolution of the AID/APOBEC family of polynucleotide (deoxy)cytidine deaminases. Mol Biol Evol 2005; 22:367 - 377; PMID: 15496550; http://dx.doi.org/10.1093/molbev/msi026
- LaRue RS, Jónsson SR, Silverstein KAT, Lajoie M, Bertrand D, El-Mabrouk N, et al. The artiodactyls APOBEC3 innate immune repertoire shows evidence for a multi-functional domain organization that existed in the ancestor of placental mammals. BMC Mol Biol 2008; 9:104; PMID: 19017397; http://dx.doi.org/10.1186/1471-2199-9-104
- Fujino T, Navaratnam N, Jarmuz A, Von Haeseler A, Scott J. C®U editing of apolipoprotein B mRNA in marsupials: identification and characterization of APOBEC-1 from the American opossum Monodelphus domestica. Nucleic Acids Res 1999; 27:2662 - 2671; PMID: 10373583; http://dx.doi.org/10.1093/nar/27.13.2662
- Severi F, Chicca A, Conticello SG. Analysis of reptilian APOBEC1 suggests that RNA editing may not be its ancestral function. Mol Biol Evol 2011; 28:1125 - 1129; PMID: 21172829; http://dx.doi.org/10.1093/molbev/msq338
- Jarmuz A, Chester A, Bayliss J, Gisbourne J, Dunham I, Scott J, et al. An anthropoid-specific locus of orphan C to U RNA-editing enzymes on chromosome 22. Genomics 2002; 79:285 - 296; PMID: 11863358; http://dx.doi.org/10.1006/geno.2002.6718
- Zhang J, Webb DM. Rapid evolution of primate antiviral enzyme APOBEC3G. Hum Mol Genet 2004; 13:1785 - 1791; PMID: 15198990; http://dx.doi.org/10.1093/hmg/ddh183
- LaRue RS, Andrésdóttir V, Blanchard Y, Conticello SG, Derse D, Emerman M, et al. Guigelines for naming nonprimate APOBEC3 genes and proteins. J Virol 2009; 83:494 - 497; PMID: 18987154; http://dx.doi.org/10.1128/JVI.01976-08