Abstract
Multiple domains of chromosomes are associated with the nuclear envelope (NE) in interphase. The association between chromosomes and the NE is involved in a variety of chromosomal reactions, such as gene expression and DNA repair. However, efficient chromosome movements are required for the fidelity of chromosome segregation in mitosis. Most higher eukaryotes perform open mitosis, in which the NE is broken down, enabling chromosomes to be released from the NE as well as spindle microtubules to access to kinetochores. By contrast, lower eukaryotes, such as Schizosaccharomyces pombe, perform closed mitosis, during which NE breakdown does not occur. In S. pombe, telomeres are tethered to the NE in interphase. Phosphorylation of the telomere-binding protein Rap1 at M phase promotes transient dissociation of telomeres from the NE, facilitating the faithful chromosome segregation. These findings imply a common mechanism for genome stability via the dissociation of chromosomes from the NE in eukaryotic mitosis.
S. pombe Telomeres Are Dissociated from the NE at M Phase
Telomeres are localized at the ends of linear chromosomes and play critical roles in the genome stability. In S. pombe, a fission yeast, the Taz1 protein (a homolog of mammalian TRF1 and TRF2) directly binds to the double-stranded telomere DNA. Taz1 directly interacts with the Rap1 protein (a homolog of mammalian Rap1) and recruits it to telomeres.Citation1-Citation3 Rap1 in turn interacts with multiple partner proteins to regulate various telomere functions, such as maintenance of telomere DNA length, protection of chromosomal ends and meiotic telomere clustering ().Citation4-Citation8
Figure 1.S. pombe telomeres are dissociated from the NE at M phase. (A) Telomere-binding proteins and their functions. (B) Telomeres are moderately clustered and are tethered to the NE in interphase. After entry into mitosis, telomeres are dissociated from the NE, and telomere clustering breaks down. For simplicity, only two chromosomes are shown.
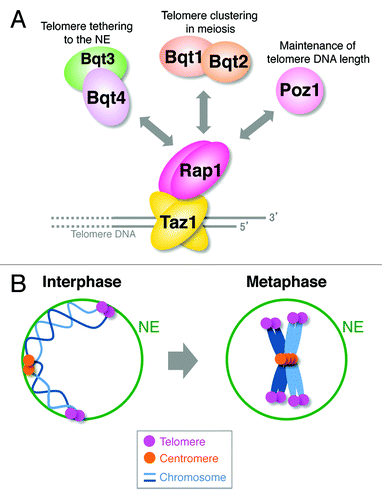
In S. pombe, telomeres moderately cluster themselves and continuously move inside the nucleus in interphase, keeping their positions near the NE ().Citation9 Telomeres are tethered to the NE via the interaction between Rap1 and the inner nuclear membrane protein Bqt4 ().Citation6 Microscopic analyses have revealed the M phase-specific movement of telomeres. Immediately after entry into mitosis, telomere clustering begins to break down, and telomeres are dissociated from the NE. At the end of mitosis, telomeres return to the NE and cluster again ().Citation9
Transient Telomere Dissociation from the NE is Required for Faithful Chromosome Segregation
To elucidate the physiological importance of the transient telomere dissociation from the NE, telomeres were forced to be tethered to the NE at M phase by expressing the fusion protein of Taz1 with the C-terminal region of Bqt4 (Bqt4∆N), which associates with the NE. The Taz1-Bqt4∆N-expressing cells displayed a higher frequency of abnormal chromosome segregation and chromosome loss than did wild-type cells. Moreover, the Taz1-Bqt4∆N-expressing cells exhibited chromosome entanglement at anaphase, suggesting that telomere dissociation from the NE is required for the efficient movement of chromosomes during mitosis and, consequently, chromosome stability.Citation9
The Telomere-Binding Protein Rap1 is Highly Phosphorylated at M Phase
Because telomeres are tethered to the NE by the interactions between the Rap1 and Bqt4 proteins in interphase, it has been speculated that the function of Rap1 changes in M phase to release telomeres from the NE. In fact, extensive band shifts of the Rap1 protein were observed at M phase in western analyses, whereas no band shift was observed for Taz1 (). The band shifts of Rap1 were primarily due to its phosphorylation at Ser213, Thr378, Ser422, Ser456 and Ser513; the timing of each phosphorylation during the cell cycle varies from site to site ().Citation9
Figure 2. Rap1 is highly phosphorylated at M phase. (A) Band shifts of the Rap1 protein in M phase. Strain JK3332 (taz1–3HA nda3-KM311) was grown in YPD medium at 32°C to mid-log phase (0 h) and gradually arrested in early M phase by a temperature shift to 20°C (4–12 h). The whole-cell extracts at each time point were analyzed by immunoblotting with anti-Rap1 antibodies, anti-HA antibodies (for Taz1-HA), anti-Cdc13 (Cyclin-B) antibodies and anti-PSTAIR antibodies (for Cdc2). (B) Schematic illustration of the identified phosphorylation sites in Rap1. BRCT, BRACA1 C terminus; Myb, Myb-related HLH motif. C, constant during the cell cycle; E–M, early to mid M phase; L, late M phase. Thr378, Ser422 and Ser513 are phosphorylated by Cdc2.
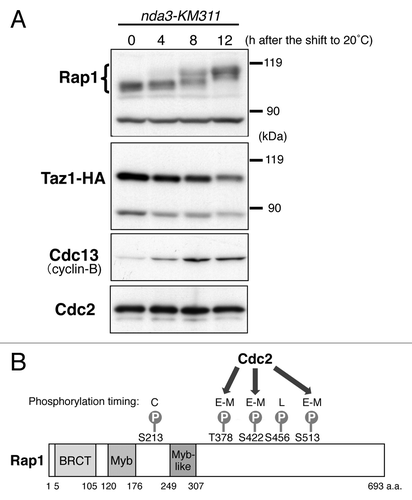
Among these phosphorylations, the phosphorylations at Thr378, Ser422 and Ser513 were observed during early to middle stages of mitosis, when Cdc2, a major mitotic kinase in S. pombe, is highly active. By contrast, these phosphorylations were barely detectable when Cdc2 was inactivated. Moreover, these phosphorylations were completely abolished when the proline residues following the three phosphorylation residues were mutated to alanine, eliminating the consensus target sequence (Ser/Thr-Pro) of Cdc2. Furthermore, p13suc1 bead-associated kinases (among which the major kinase is Cdc2) efficiently phosphorylated these three residues in vitro. Taken together, these results strongly suggest that Cdc2 phosphorylates these three residues.Citation9 However, the identity of the kinase(s) that phosphorylate Ser213 and Ser456 of Rap1 is unknown ().
Phosphorylation of Rap1 Promotes Telomere Dissociation from the NE
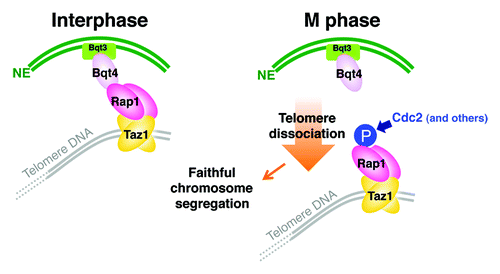
Phosphorylation of Rap1 inhibits the interaction between Rap1 and Bqt4. Furthermore, substitution of amino acid residues at the five phosphorylation sites with alanine decreases the distances between the telomeres and the NE. By contrast, substitution with phospho-mimic amino acid residues (aspartic acid or glutamic acid) increases the distance, as in the case of the rap1+ or bqt4+ deletion. The phosphorylation of Rap1 specifically influences the interaction between Rap1 and Bqt4, but not the interactions between Rap1 and Taz1, Poz1 and Bqt1-Bqt2. Consistently, alanine- or phospho-mimic mutations of rap1+ at the phosphorylation sites do not exhibit any defect in telomere DNA length or the progression of meiosis (). Moreover, among the five phosphorylation sites of Rap1, the phosphorylation at Ser513 is most influential on the Rap1-Bqt4 interaction and, consequently, the distance between the telomeres and the NE.Citation9 These findings indicate that the activation of Cdc2 kinase at early M phase is the major trigger for the release of telomeres from the NE via Rap1 phosphorylation, which reduces the association between Rap1 and Bqt4 ().
Figure 3. Model for regulation of telomere localization during the cell cycle in S. pombe. Telomeres are tethered to the NE via the Taz1-Rap1-Bqt4-Bqt3 association in interphase. During mitosis, Rap1 is phosphorylated. This phosphorylation, primarily by Cdc2, promotes the detachment of Rap1 from Bqt4 and thereby telomere dissociation from the NE. Transient telomere dissociation from the NE in mitosis is required for faithful chromosome segregation.
Dissociation of Other Chromosomal Domains from the NE at M Phase
Deletion of Rap1 or Bqt4 results in the detachment of “telomere ends” from the NE; however, most of the telomere ends in the rap1∆ or bqt4∆ strains are located relatively close to the NE,Citation9 suggesting that there are other mechanisms by which the telomere ends are maintained in the vicinity of the NE in interphase. Steglich and colleagues reported that Man1, an inner nuclear membrane protein, preferentially binds to the chromosomal regions around the sub-telomeres and is involved in their localization at the nuclear periphery. Furthermore, genes with low expression levels and heterochromatin loci are generally located at the nuclear periphery.Citation10 Thus, multiple chromosomal domains are located at the nuclear periphery in S. pombe as in other eukaryotes.Citation11-Citation14 This may be why the telomere ends do not freely float in the nucleus, even in the absence of Rap1 or Bqt4. It is highly possible that mitosis-specific dissociation from the NE occurs not only at telomeres but also at other chromosomal regions. In fact, S. pombe centromeres are clustered toward the spindle pole body (SPB) at the nuclear periphery in interphase.Citation15 Upon entry into mitosis, centromeres are dissociated from the SPB and from the NE and are relocated at the center of the nucleus. However, how these other chromosomal regions at the nuclear periphery become released from the NE in mitosis remains unclear.
Telomere-NE Dissociation and Telomere Declustering for the Progression of Mitosis
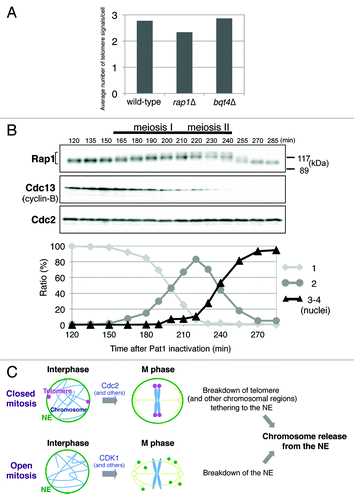
In S. pombe, telomeres continuously move inside the nucleus in interphase. Therefore, it is highly possible that the long chromatin fibers are entangled with each other because a large number of chromosomal domains are associated with the NE. For the normal activation of spindle assembly checkpoint, which detects the tension between the sister kinetochores, each chromosome must be aligned at the center of the nucleus at metaphase without any entanglement or tension between the kinetochores and the NE. Thus, dynamism of telomeres in mitosis, i.e., detachment from the NE and declustering, may facilitate the resolution of chromatin fibers before metaphase. In fact, entanglement of chromosomes has been observed at anaphase when telomeres are artificially tethered to the NE in mitosis.Citation9 Although Rap1 is crucial for the “meiotic” telomere clustering toward SPB,Citation2,Citation3 telomeres are normally clustered in interphase in rap1 or bqt4 deletion strains (), indicating that the other proteins are involved in telomere clustering in the mitotic cell cycle; however, how telomere clustering and declustering are regulated is largely unknown.
Figure 4. Conservation of chromosome release from the NE in eukaryotic cell division. (A) Mitotic telomere clustering in rap1∆ and bqt4∆ cells. Strains JK81 (wild-type, taz1-mCherry), JP836 (rap1∆) and JP842 (bqt4∆) were grown in YES medium at 30°C, and the numbers of Taz1-mCherry signals per cell were counted. More than 100 cells in G2 phase were analyzed for each strain. Note that S. pombe has three chromosomes and six telomeres. (B) Modification of the Rap1 protein in meiosis. Strain JP791 (h-/h-pat1–114/pat1–114 mat-Pc) was grown in YE medium to mid-log phase at 25°C and then transferred to EMM-N medium (supplemented with 1% glucose) at 25°C. After 6.5 h of incubation (G1 arrest), the temperature was shifted to 34°C to inactivate Pat1. Samples were taken every 10–15 min after 120 min of incubation at 34°C. The percentage of cells with 1, 2 or 3 –4 nuclei was determined by counting > 200 cells stained with DAPI (4',6-diamidino-2-phenylindole). The whole-cell extracts were analyzed by immunoblotting with anti-Rap1 antibodies, anti-Cdc13 (Cyclin-B) antibodies and anti-PSTAIR antibodies for Cdc2 (loading control). (C) Models for the regulation of closed and open mitoses. The breakdown of telomere tethering to the NE is triggered primarily by Cdc2 in closed mitosis in S. pombe. Dissociation of the other chromosomal regions from the NE may also be regulated by Cdc2. By contrast, breakdown of the NE triggered by CDK1 and other mitotic kinases occurs in open mitosis. Both induce the release of chromosomes from the NE, which likely leads to accurate chromosome segregation.
Chromosome Release from the NE: The Universal Principle of Eukaryotic Mitosis Involving Phosphorylation Events
A budding yeast, Saccharomyces cerevisiae, also performs closed mitosis. In this organism, telomeres are attached to NE through multiple pathways that involve telomere-binding proteins, Sir4 and yKu80, and inner nuclear membrane proteins, Esc1 and Mps3.Citation13,Citation16 The redundant mechanisms of telomere tethering to NE imply that telomere localization is important for cellular activities. In fact, recombination between sub-telomeres is repressed, and DNA repair in sub-telomeric region is regulated by telomere positioning at nuclear periphery.Citation17,Citation18 It has been shown that yKu80-dependent telomere anchoring to the NE is switched off after DNA replication,Citation19 suggesting that a mechanism similar to that in S. pombe mitosis exists in S. cerevisiae to facilitate the faithful chromosome segregation in mitosis.
In mouse meiosis, telomeres are clustered to a limited area at the nuclear periphery at the zygotene stage as in S. pombe meiosis, and dissociation of the telomeres from the NE occurs in later stages.Citation20,Citation21 Thus, the regulatory mechanism of telomere dissociation from the NE is most likely not specific to closed mitosis, that is, open and closed meioses may also follow a mechanism similar to that of closed mitosis to release telomeres from the NE. Consistent with this suggestion, S. pombe Rap1 is highly modified during meiosis I and II ().
Telomere dissociation from the NE in S. pombe mitosis is promoted by Rap1 phosphorylation, which is primarily dependent on Cdc2. In open mitosis, phosphorylation of lamin, some components of the nuclear pore complex (NPC) and several inner nuclear membrane proteins by CDK1 and other mitotic kinases is important for disassembly of the NE and NPC.Citation22 In addition, the mitotic kinase of the VRK (vaccinia-related kinase) family phosphorylates the chromatin-binding protein BAF (barrier-to-autointegration factor), which links chromosomes with the NE, to reduce the affinity of BAF for chromosomes.Citation23 Therefore, phosphorylation of proteins at the nuclear periphery appears to be a key and universal mechanism to release chromosomes from the NE in both open and closed mitoses. As described above, a large number of chromosomal regions in addition to telomeres are located near the NE in S. pombe interphase. It will be of interest to determine whether mitotic dissociation of the other chromosomal regions from the NE is also regulated by phosphorylation of the chromatin-associated proteins. Taken together, in closed mitosis, cells show breakdown of tethering of the telomere (and possibly other chromosomal regions) to the NE, while chromosomes become free in open mitosis by breakdown of the NE. In both cases, chromosomes are transiently released from the NE, which may contribute to efficient chromosome movements and, consequently, fidelity of chromosome segregation ().
Acknowledgments
I thank Ayumu Yamamoto for the S. pombe strain and Kayoko Tanaka and Akira Yamashita for technical advice. This work was supported by the Osaka University Life Science Young Independent Researcher Support Program of JST, Grants-in-Aid for Scientific Research (KAKENHI), the Astellas Foundation for Metabolic Disorders, the Takeda Science Foundation, the Sumitomo Foundation and the Novartis Foundation for the Promotion of Science.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- Cooper JP, Nimmo ER, Allshire RC, Cech TR. Regulation of telomere length and function by a Myb-domain protein in fission yeast. Nature 1997; 385:744 - 7; http://dx.doi.org/10.1038/385744a0; PMID: 9034194
- Chikashige Y, Hiraoka Y. Telomere binding of the Rap1 protein is required for meiosis in fission yeast. Curr Biol 2001; 11:1618 - 23; http://dx.doi.org/10.1016/S0960-9822(01)00457-2; PMID: 11676924
- Kanoh J, Ishikawa F. spRap1 and spRif1, recruited to telomeres by Taz1, are essential for telomere function in fission yeast. Curr Biol 2001; 11:1624 - 30; http://dx.doi.org/10.1016/S0960-9822(01)00503-6; PMID: 11676925
- Miller KM, Ferreira MG, Cooper JP. Taz1, Rap1 and Rif1 act both interdependently and independently to maintain telomeres. EMBO J 2005; 24:3128 - 35; http://dx.doi.org/10.1038/sj.emboj.7600779; PMID: 16096639
- Chikashige Y, Tsutsumi C, Yamane M, Okamasa K, Haraguchi T, Hiraoka Y. Meiotic proteins Bqt1 and Bqt2 tether telomeres to form the bouquet arrangement of chromosomes. Cell 2006; 125:59 - 69; http://dx.doi.org/10.1016/j.cell.2006.01.048; PMID: 16615890
- Chikashige Y, Yamane M, Okamasa K, Tsutsumi C, Kojidani T, Sato M, et al. Membrane proteins Bqt3 and -4 anchor telomeres to the nuclear envelope to ensure chromosomal bouquet formation. J Cell Biol 2009; 187:413 - 27; http://dx.doi.org/10.1083/jcb.200902122; PMID: 19948484
- Miyoshi T, Kanoh J, Saito M, Ishikawa F. Fission yeast Pot1-Tpp1 protects telomeres and regulates telomere length. Science 2008; 320:1341 - 4; http://dx.doi.org/10.1126/science.1154819; PMID: 18535244
- Fujita I, Tanaka M, Kanoh J. Identification of the functional domains of the telomere protein Rap1 in Schizosaccharomyces pombe.. PLoS One 2012; 7:e49151; http://dx.doi.org/10.1371/journal.pone.0049151; PMID: 23133674
- Fujita I, Nishihara Y, Tanaka M, Tsujii H, Chikashige Y, Watanabe Y, et al. Telomere-nuclear envelope dissociation promoted by Rap1 phosphorylation ensures faithful chromosome segregation. Curr Biol 2012; 22:1932 - 7; http://dx.doi.org/10.1016/j.cub.2012.08.019; PMID: 22959349
- Steglich B, Filion GJ, van Steensel B, Ekwall K. The inner nuclear membrane proteins Man1 and Ima1 link to two different types of chromatin at the nuclear periphery in S. pombe.. Nucleus 2012; 3:77 - 87; http://dx.doi.org/10.4161/nucl.18825; PMID: 22156748
- Weierich C, Brero A, Stein S, von Hase J, Cremer C, Cremer T, et al. Three-dimensional arrangements of centromeres and telomeres in nuclei of human and murine lymphocytes. Chromosome Res 2003; 11:485 - 502; http://dx.doi.org/10.1023/A:1025016828544; PMID: 12971724
- Bolzer A, Kreth G, Solovei I, Koehler D, Saracoglu K, Fauth C, et al. Three-dimensional maps of all chromosomes in human male fibroblast nuclei and prometaphase rosettes. PLoS Biol 2005; 3:e157; http://dx.doi.org/10.1371/journal.pbio.0030157; PMID: 15839726
- Mekhail K, Moazed D. The nuclear envelope in genome organization, expression and stability. Nat Rev Mol Cell Biol 2010; 11:317 - 28; http://dx.doi.org/10.1038/nrm2894; PMID: 20414256
- Taddei A, Gasser SM. Structure and function in the budding yeast nucleus. Genetics 2012; 192:107 - 29; http://dx.doi.org/10.1534/genetics.112.140608; PMID: 22964839
- Takahashi K, Murakami S, Chikashige Y, Funabiki H, Niwa O, Yanagida M. A low copy number central sequence with strict symmetry and unusual chromatin structure in fission yeast centromere. Mol Biol Cell 1992; 3:819 - 35; PMID: 1515677
- Taddei A, Schober H, Gasser SM. The budding yeast nucleus. Cold Spring Harb Perspect Biol 2010; 2:a000612; http://dx.doi.org/10.1101/cshperspect.a000612; PMID: 20554704
- Therizols P, Fairhead C, Cabal GG, Genovesio A, Olivo-Marin JC, Dujon B, et al. Telomere tethering at the nuclear periphery is essential for efficient DNA double strand break repair in subtelomeric region. J Cell Biol 2006; 172:189 - 99; http://dx.doi.org/10.1083/jcb.200505159; PMID: 16418532
- Schober H, Ferreira H, Kalck V, Gehlen LR, Gasser SM. Yeast telomerase and the SUN domain protein Mps3 anchor telomeres and repress subtelomeric recombination. Genes Dev 2009; 23:928 - 38; http://dx.doi.org/10.1101/gad.1787509; PMID: 19390087
- Ebrahimi H, Donaldson AD. Release of yeast telomeres from the nuclear periphery is triggered by replication and maintained by suppression of Ku-mediated anchoring. Genes Dev 2008; 22:3363 - 74; http://dx.doi.org/10.1101/gad.486208; PMID: 19056887
- Chikashige Y, Ding DQ, Funabiki H, Haraguchi T, Mashiko S, Yanagida M, et al. Telomere-led premeiotic chromosome movement in fission yeast. Science 1994; 264:270 - 3; http://dx.doi.org/10.1126/science.8146661; PMID: 8146661
- Scherthan H, Weich S, Schwegler H, Heyting C, Härle M, Cremer T. Centromere and telomere movements during early meiotic prophase of mouse and man are associated with the onset of chromosome pairing. J Cell Biol 1996; 134:1109 - 25; http://dx.doi.org/10.1083/jcb.134.5.1109; PMID: 8794855
- Güttinger S, Laurell E, Kutay U. Orchestrating nuclear envelope disassembly and reassembly during mitosis. Nat Rev Mol Cell Biol 2009; 10:178 - 91; http://dx.doi.org/10.1038/nrm2641; PMID: 19234477
- Gorjánácz M, Klerkx EP, Galy V, Santarella R, López-Iglesias C, Askjaer P, et al. Caenorhabditis elegans BAF-1 and its kinase VRK-1 participate directly in post-mitotic nuclear envelope assembly. EMBO J 2007; 26:132 - 43; http://dx.doi.org/10.1038/sj.emboj.7601470; PMID: 17170708