Abstract
Many if not most proteins can, under certain conditions, change cellular compartments, such as, for example, shuttling from the cytoplasm to the nucleus. Thus, many proteins may exert functions in various and very different subcellular locations, depending on the signaling context. A large amount of actin regulatory proteins has been detected in the mammalian cell nucleus, although their potential roles are much debated and are just beginning to emerge. Recently, members of the formin family of actin nucleators were also reported to dynamically localize to the nuclear environment. Here we discuss our findings that specific diaphanous-related formins can promote nuclear actin assembly in a signal-dependent manner.
Introduction
An enormous amount of literature exists that describes an array of cellular and molecular functions for diaphanous-related formins mostly involving actin-based membrane protrusions, migration, contractility, adhesion, cytokinesis, and microtubule regulation (reviewed in refs. Citation1, Citation2). In addition, a few studies suggested that some formins may also be detected in the nuclear compartment. Indirect hints came from copurifications of mDia1 with proteins of predominantly nuclear functions such as exportin 6 or the transcriptional regulator HAN11.Citation3,Citation4 The formin FHOD1 can be cleaved by caspase-3 generating a fragment with strong nuclear localization.Citation5 Convincing evidence for cytoplasmic-nuclear shuttling of a formin came from Miki et al. demonstrating CRM1-dependent export of endogenous mDia2.Citation6 However, whether these nuclear localizations involve any cellular functions or whether formins may be active in the nucleus remained unclear. Moreover, formins potently promote actin nucleation and filament assembly, a process that previously has not been described to occur in a somatic cell nucleus. Recently, that has changed by the first demonstrations of nuclear actin polymers in living cells (also reviewed in refs. Citation7–Citation9).
Here, after a short summary of the current view on nuclear actin dynamics, we briefly discuss the potential roles of nuclear formin regulation and activity for actin nucleation and MAL/SRF transcriptional function.
Nuclear Actin Dynamics
By altering the concentration of signal-competent G-actin or by supplying the huge amount of cellular processes, which rely on the formation of actin filaments, it’s the dynamic assembly and disassembly of actin polymers, which attributes to virtually all of its biological impact. In light of an ever-expanding body of evidence reinforcing the biological significance of actin inside the mammalian nucleus,Citation7,Citation10,Citation11 we however only recently began to understand the dynamic nature of nuclear actin.
FRAP experiments demonstrated a dynamic exchange of GFP-actin monomers across the nuclear envelopeCitation12 and provided the first hints of a subpopulation of nuclear actin residing in a stable, less diffusible state.Citation13 Accordingly, our recently published work and the study of Belin and collegues demonstrated the first direct visualizations of nuclear actin polymers.Citation14,Citation15 Together, these findings strongly suggest that at least a certain pool of nuclear actin exists in a dynamic equilibrium between G- and F-actin resembling the treadmilling of cytosolic actin. By applying in-vitro actin assembly assays using nuclear extracts, we could show that the nucleoplasm possesses a basal degree of actin polymerizing activity. In agreement with this, nuclear F-actin structures of submicron-length have been detected in the nuclei of non-stimulated cells using the nuclear F-actin probe Utr230-EN.Citation15 These findings raise important questions: how, and if so, to what extent, can nuclear actin turnover be altered to eventually become instrumental in controlling certain aspects of cell behavior?
During our studies we obtained several results indicating that the state of nuclear actin polymerization is indeed subject to alterations. In addition, we provide several lines of evidence for a critical contribution of Diaphanous-related formins, namely mDia1 and mDia2, in controlling nuclear actin dynamics. First, the siRNA-mediated silencing of either mDia1 or mDia2 resulted in a considerable decrease of basal actin polymerization activity of nuclear extracts. Second, nuclear expression of the mDia2-DAD domain, which is known to promote the activity of endogenous mDia, sufficiently shifts nuclear actin dynamics toward polymerization giving rise to nuclear actin filaments, which become detectable in living cells. Third, the acute stimulation of cellular actin assembly using serum or the serum component LPA (lysophosphatidic acid) triggers a transient burst of actin polymerization inside the nuclear compartment (). Of note, this nuclear polymerization response is dependent on the activity of mDia formins and appears, at least in the case of NIH3T3 cells, even strong enough to promote the transient formation of phalloidin-sensitive nuclear actin filaments.
Figure 1. Signal-responsive nuclear actin dynamics. Live NIH3T3 cells expressing the actin probe LifeAct-GFP-NLS were monitored before and during stimulation with 20 µM lysophosphatidic acid (LPA). Prior to analysis, cells were transiently transfected with a plasmid encoding LifeAct-GFP-NLS and kept in serum-free medium for 24 h. Confocal microscopic images (1 frame every 10 s) reveal the distribution of LifeAct-GFP-NLS at indicated time points. Note that LPA-stimulation triggers an immediate and transient formation of nuclear actin filaments, which become visible by the decoration with LifeAct-GFP-NLS. Scale bar, 10 µm.
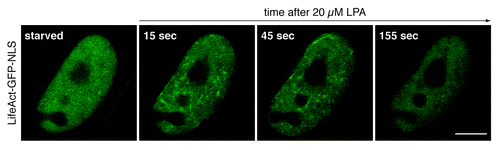
Thus, the nature of nuclear actin appears to be much more dynamic than previously thought. Overall, a picture emerges in which not only the concentration of nuclear actin underlies tight control but also its polymerization state, which relies on the activity of actin-regulatory factors present inside the nucleus. Finally both, the dynamic communication of actin monomers across the nuclear envelope, as well as the existence of a treadmill-competent pool, may act in concert to sustainably equip nuclear actin for a substantial expansion of its biological properties.
Regulation of Nuclear Actin Turnover by Formins
The importance of mDia formins in defining the polymerization state of nuclear actin is further underscored by our findings that neither a depletion of the formin FHOD1 using siRNA nor chemical inhibition of the Arp2/3-complex affected the actin-polymerizing activity of nuclear extracts. Whereas a huge body of literature exists that addresses the regulation and function of mDia-formins in rearranging cytosolic actin,Citation1,Citation2 the mechanisms at work to control nuclear mDia-function remain enigmatic. Our observation of a very rapid and short-lived stimulation of mDia-dependent nuclear actin assembly upon serum-addition argues for a tight regulation of nuclear formin activity. In an attempt to transfer the present knowledge on cytosolic formin function to the nucleus, we would like to highlight three potential aspects of nuclear formin regulation ().
Figure 2. Cartoon illustrating potential mechanisms involved in the control of formin-mediated assembly of nuclear actin. For details see text.
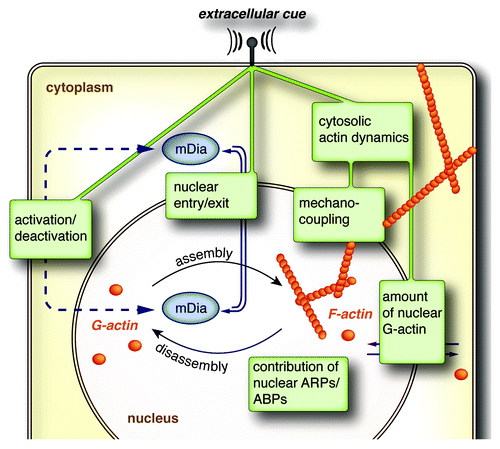
(1) Nuclear recruitment of formins. So far, the underlying mechanisms of nuclear formin transport have only really been characterized for mDia2.Citation6 Full-length mDia2 is recognized by importin-α, which binds to a bipartite nuclear localization signal (NLS) involving the amino acids Lys35 and Arg36. Nuclear export of mDia2 is facilitated by the export receptor CRM1 (exportin 1), which can be targeted by the drug Leptomycin B (LMB). Under steady-state conditions mDia2 is mainly found in the cytoplasm arguing for a highly dynamic nucleo-cytosolic shuttling, which is further reflected by an immediate and almost complete nuclear accumulation within minutes after LMB-treatment. In contrast to mDia2, the localization of the related formins mDia1 and mDia3 appears insensitive to LMB-treatment.Citation6 However, this observation does not exclude the possibility of alternative nuclear export mechanisms acting independent of CRM1. Interestingly, endogenous mDia1 was found to co-precipitate with exportin 6, which appears critically involved in nuclear export of actin and profilin-actin complexes.Citation4 Moreover, several reports indicate the presence of a putative NLS in the C-terminus of mDia1, which is consistent with a predominant nuclear localization of a C-terminal fragment of mDia1 comprising its FH1, FH2, and DAD domains.Citation16 Whether this NLS may also function in full-length mDia1 remains to be tested. In this regard the mDia1 N-terminus could either prevent nuclear entry or antagonize nuclear localization of mDia1 by promoting its nuclear export. Overall, the shuttling behavior of mDia2 underscores the need for a careful reassessment before ruling out an eventually substantial nuclear transport of a given formin, despite its presumable cytosolic localization. Alterations in shuttling dynamics might serve as a powerful tool to adjust the amount of nuclear formins in a signal-dependent manner. However, available techniques allowing for a detailed analysis of formin nuclear transport kinetics are hampered by the limited value of studying fluorescently-labeled and ectopically expressed proteins, which do not necessarily reflect the behavior of their endogenous counterparts (unpublished observation on GFP-labeled mDia2). At present we can only speculate about the amount of nuclear formins necessary to account for nuclear actin assembly. Moreover, nuclear formins might preferentially reside in close proximity to the inner membrane of the nuclear envelope further complicating a reliable assessment of their nuclear localization. Of note, both mDia1 and mDia2 were shown to intrinsically attach to lipid membranes,Citation17,Citation18 which in the case of mDia2 can be mediated by a basic stretch of amino acids located right next to the NLS. In this regard, it will become an interesting future perspective to define the spatial origin of formin-mediated nuclear actin polymerization.
(2) Activation of nuclear formins. Diaphanous-related formins are characterized by an autoinhibited conformation, in which interactions between their N-terminal FH3 domains and their C-terminal DAD regions physically prevent the ability to polymerize actin under resting conditions. Therefore, the nuclear activity of formins not only relies on their recruitment to the nucleus but also claims for a release of autoinhibition. We don’t know at present whether the activity-state of formins affects nuclear import, as activation can, at least in principle, occur in the cytoplasm (prior to nuclear import), as well as in the nucleus itself.Citation7 Since the majority of mDia formins is believed to reside in an autoinhibited state under resting conditions, the rapid and almost complete nuclear accumulation of mDia2 upon CRM1 inhibition suggests nuclear entry of autoinhibited mDia2. However, it remains to be tested whether the structurally open conformation interferes with its nuclear import. Accordingly, only the failure of endogenous active mDia2 to enter the nucleus (which might for example occur due to conformational hindrance or an immediate association to cytosolic actin filament ends) would require a specialized nuclear mechanism of activation. On the other hand, nuclear activity of formins could be primarily defined by their activation state in the cytoplasm, which would argue for a more or less passive transduction of formin activity toward the nuclear compartment. During our studies we made use of a known competition mechanism, in which expression of the DAD peptide competes with the autoinhibitory interactions of endogenous mDia.Citation19 Whereas nuclear expression of the DAD peptide allowed us to selectively promote the activity of endogenous nuclear mDia it remains elusive whether the DAD is indeed sufficient to release formin autoinhibition or whether its activity primarily arises by stabilizing an already pre-activated conformation. Therefore, it will be an important future task to first ensure the true nuclear origin of formin activation before aiming to transfer the elaborated cellular system of regulatory mechanisms, known to participate in the spatial and temporal control of cytosolic formins, to the nucleus.
The classical view of formin activation involves the binding of active Rho-GTPases to their GBD (GTPase binding domain), which due to its close proximity to the FH3 domain results in a displacement of the DAD. Whether Rho-GTPases exert similar activities inside the nucleus remains to be proven. Despite their small size (<24 kD), which would at least in theory allow for a passive diffusion through the nuclear pore complex and the presence of NLS sequences in the C-termini of many Ras- and Rho-family GTPases,Citation20 biochemical fractionations suggest a comparatively low amount of endogenous Rho-GTPases present in the nucleus.Citation21,Citation22 However, there is also mounting evidence for additional cues being involved in controlling both the subcellular recruitment and/or the activation state of certain formins. In particular phosphorylation of residues within the DAD emerges as an important GTPase-independent mode of formin activation,Citation1 and some of the involved kinases such as Aurora B or Casein-Kinase 2 (CK2) are also abundant in the nucleus. Moreover, not only the list of formins are subject to post-translational modifications, but also the number of formin-interacting proteins steadily expands, which further adds complexity to potential activation mechanisms underlying formin activity inside the nucleus.
Once activated, formins may affect nuclear actin dynamic by either de-novo nucleation of actin filaments or by their ability to processively elongate pre-existing filament barbed ends. Whereas the elongation activity of formins appears to be an intrinsic feature of their biochemical properties, the precise mechanisms underlying the initiation of filament growth are still under debate. Several studies argue for a critical involvement of nucleation promoting factors (NPF), which cooperate with formins to facilitate efficient actin nucleation. Examples are provided by the NPF-formin pair Bud6-Bni1 in yeast, the collaborative activities of Spire and Formin-2 (FMN2) and the recently identified dependence of mDia1 on APC.Citation1 Currently, we don’t know to what extent the nuclear function of formins is attributed to their nucleation activity but the presence of several NPFs (e.g., APC, Arp2/3, or JMY) in the nucleus provides a promising future perspective on potential collaborative functionalities.
(3) Termination of nuclear formin activity. The absence of readily detectable nuclear actin filaments in non-stimulated cells as well as the transient nature of serum-stimulated nuclear actin assembly not only suggests a polymerization on demand but furthers the presence of efficient nuclear control mechanisms to restrict the duration and actin-polymerization rates of nuclear formins. One nearby model of limiting the action of nuclear formins would be given by a controlled nuclear export of activated formins. But whether the activated state of formins is able to exit the nucleus remains to be tested.
In vitro observations indicate that once activated, formins persist on the progressively growing ends of actin filaments for more than ten minutes,Citation23 which in vivo would most likely interfere with nuclear export. Alternatively, the nucleoplasm might harbor additional regulatory factors, which may act by promoting a displacement of activated formins from filament ends. Similar mechanisms have already been described for Capping protein, which competes with mDia1 for binding to the barbed ends of cytosolic actin filaments and the formin-binding protein srGAP2, which was shown to limit the duration of FMNL1 activity upon Rac-dependent activation.Citation1 Given the overall huge number of actin-binding (ABPs) and actin-related proteins (ARPs) present in the nucleoplasm,Citation10,Citation11 it will remain a future challenge to test for their potential impact in coordinating nuclear formin function.
Cytosolic Impact on Nuclear Actin Dynamic
Aiming for the nuclear compartment, signals have to overcome the nuclear envelope, which ensures a considerable confinement of the nucleoplasm from the cytosol. This raises the question of how environmental cues, sensed by cell surface receptors, may become integrated and transmitted toward the nuclear interior to ultimately affect nuclear actin dynamics. Despite being just a passive relay station, the cytoplasm and especially cytosolic actin dynamics are likely to participate more actively in defining the polymerization state of nuclear actin. The demonstration of a dynamic communication between nuclear and cytosolic actin monomers suggests a model in which the concentration of nuclear actin reflects its cytosolic turnover.Citation12 However, whether the underlying shuttling kinetics of actin are sufficient to transduce even short-term alterations in its cytosolic turnover to the nuclear compartment remains to be tested. On the other hand, the cytosolic actin network might be much more directly coupled to its nuclear descendant through LINC (linkers of the nucleoskeleton to the cytoskeleton) protein complexes. LINC complexes span the nuclear envelope (NE) and arise by the physical interactions of transmembrane proteins of the outer NE-membrane, namely nesprins, and SUN-proteins, which in turn reside in the inner membrane of the NE.Citation24 The ability of SUN-proteins to associate with the nuclear lamina, together with the capability of nesprin1/2 to bind actin filaments enables LINC complexes to physically connect the nuclear interior to the actin cytoskeleton. Although we don’t know at present whether the nuclear lamina associates with nuclear actin it is worth noticing that both A-type and B-type lamins as well as emerin, a protein of the inner nuclear membrane, were reported to directly interact with actin in vitro.Citation25,Citation26 Hence, given its location at the interface between nuclear and cytosolic actin as well as its key function in mechano-signaling to the nucleus it is tempting to speculate about an impact of LINC-mediated transduction of tensional forces originating from cytosolic actin rearrangements on nuclear actin dynamics. Moreover, physical forces recently emerged as a novel mechanism to control the actin assembly rates of formins, as shown for mDia1Citation27 and the yeast formin Bni1p.Citation28 Such tension-based activation mechanism would further not require any additional nuclear factor for release of formin autoinhibition and would also circumvent the requirement for nuclear import of an already preactivated mDia.
Overall, the autonomy of nuclear actin dynamics remains elusive, and we believe that care should be taken to consider nuclear actin polymerization as an isolated cellular response. During our studies, we could show that solely the stimulation of nuclear formin activity, which we achieved by either nuclear targeting or photoactivation of the DAD peptide, is sufficient to promote nuclear actin filament formation.Citation14 Whereas these experiments demonstrate that the nuclear actin polymerization machinery can, at least in principle, be selectively targeted, they do not address the presence of endogenous signaling pathways dedicated to the control of nuclear actin dynamics. Instead, the regulation of the transcriptional co-activator MAL (also known as MRTF-A and MKL1) points toward an elaborated interplay between nuclear and cytosolic actin dynamics.Citation29
Interestingly, the activity of MAL is inhibited by monomeric actin, which binds to its RPEL domain in both cellular compartments. Binding of up to five actin monomers enables MAL to precisely sense the amount of cellular G-actin, resulting in a continuous shuttling through the nuclear compartment under resting conditions.Citation30 Whereas nuclear actin monomers promote MAL nuclear export, the excessive amount of cytosolic G-actin impairs its nuclear entry.Citation29,Citation31 Hence, MAL activity requires efficient disruption of actin-MAL complexes in both compartments, which is believed to occur as an indirect consequence of a polymerization-induced depletion of cellular actin monomers. In this regard, our work uncovered a so-far neglected direct contribution of nuclear actin polymers, which on its own appears sufficient to promote MAL activationCitation14 but which most likely occurs as part of a global cellular polymerization response converging in efficient control of MAL-dependent gene expression.
Conclusion
Many actin regulatory proteins such as mDia2 are found in the cytoplasm and nucleus, and actin itself dynamically shuttles between these two compartments. This suggests that the cellular cytoskeleton tightly links actin dynamics between cytoplasm and nucleus to provide a highly sensitive means of intracellular communication. Elucidating the mechanisms and consequences of formin-mediated nuclear actin assembly as well as further structural insights into its organization remain a future challenge.
Disclosure of Potential Conflicts of Interest
No potential conflict of interest was disclosed.
Acknowledgments
We thank laboratory members and colleagues for discussions. Our work is supported by the DFG (GR 2111/2-1; SFB 593).
References
- Breitsprecher D, Goode BL. Formins at a glance. J Cell Sci 2013; 126:1 - 7; http://dx.doi.org/10.1242/jcs.107250; PMID: 23516326
- Baarlink C, Brandt D, Grosse R. SnapShot: Formins. Cell 2010; 142:172 - , e1; http://dx.doi.org/10.1016/j.cell.2010.06.030; PMID: 20603022
- Stüven T, Hartmann E, Görlich D. Exportin 6: a novel nuclear export receptor that is specific for profilin.actin complexes. EMBO J 2003; 22:5928 - 40; http://dx.doi.org/10.1093/emboj/cdg565; PMID: 14592989
- Morita K, Lo Celso C, Spencer-Dene B, Zouboulis CC, Watt FM. HAN11 binds mDia1 and controls GLI1 transcriptional activity. J Dermatol Sci 2006; 44:11 - 20; http://dx.doi.org/10.1016/j.jdermsci.2006.06.001; PMID: 16887337
- Ménard I, Gervais FG, Nicholson DW, Roy S. Caspase-3 cleaves the formin-homology-domain-containing protein FHOD1 during apoptosis to generate a C-terminal fragment that is targeted to the nucleolus. Apoptosis 2006; 11:1863 - 76; http://dx.doi.org/10.1007/s10495-006-0087-8; PMID: 17013756
- Miki T, Okawa K, Sekimoto T, Yoneda Y, Watanabe S, Ishizaki T, Narumiya S. mDia2 shuttles between the nucleus and the cytoplasm through the importin-alpha/beta- and CRM1-mediated nuclear transport mechanism. J Biol Chem 2009; 284:5753 - 62; http://dx.doi.org/10.1074/jbc.M806191200; PMID: 19117945
- Treisman R. Shedding light on nuclear actin dynamics and function. Trends Biochem Sci 2013; 38:376 - 7; http://dx.doi.org/10.1016/j.tibs.2013.06.004; PMID: 23810602
- Belin BJ, Mullins RD. What we talk about when we talk about nuclear actin. Nucleus 2013; 4:291 - 7; http://dx.doi.org/10.4161/nucl.25960; PMID: 23934079
- Grosse R, Vartiainen MK. To be or not to be assembled: progressing into nuclear actin filaments. Nat Rev Mol Cell Biol 2013; 14:693 - 7; http://dx.doi.org/10.1038/nrm3681; PMID: 24088744
- Gieni RS, Hendzel MJ. Actin dynamics and functions in the interphase nucleus: moving toward an understanding of nuclear polymeric actin. Biochem Cell Biol 2009; 87:283 - 306; http://dx.doi.org/10.1139/O08-133; PMID: 19234542
- Percipalle P. Co-transcriptional nuclear actin dynamics. Nucleus 2013; 4:43 - 52; http://dx.doi.org/10.4161/nucl.22798; PMID: 23138849
- Dopie J, Skarp K-P, Rajakylä EK, Tanhuanpää K, Vartiainen MK. Active maintenance of nuclear actin by importin 9 supports transcription. Proc Natl Acad Sci U S A 2012; 109:E544 - 52; http://dx.doi.org/10.1073/pnas.1118880109; PMID: 22323606
- McDonald D, Carrero G, Andrin C, de Vries G, Hendzel MJ. Nucleoplasmic beta-actin exists in a dynamic equilibrium between low-mobility polymeric species and rapidly diffusing populations. J Cell Biol 2006; 172:541 - 52; http://dx.doi.org/10.1083/jcb.200507101; PMID: 16476775
- Baarlink C, Wang H, Grosse R. Nuclear actin network assembly by formins regulates the SRF coactivator MAL. Science 2013; 340:864 - 7; http://dx.doi.org/10.1126/science.1235038; PMID: 23558171
- Belin BJ, Cimini BA, Blackburn EH, Mullins RD. Visualization of actin filaments and monomers in somatic cell nuclei. Mol Biol Cell 2013; 24:982 - 94; http://dx.doi.org/10.1091/mbc.E12-09-0685; PMID: 23447706
- Copeland SJ, Green BJ, Burchat S, Papalia GA, Banner D, Copeland JW. The diaphanous inhibitory domain/diaphanous autoregulatory domain interaction is able to mediate heterodimerization between mDia1 and mDia2. J Biol Chem 2007; 282:30120 - 30; http://dx.doi.org/10.1074/jbc.M703834200; PMID: 17716977
- Gorelik R, Yang C, Kameswaran V, Dominguez R, Svitkina T. Mechanisms of plasma membrane targeting of formin mDia2 through its amino terminal domains. Mol Biol Cell 2011; 22:189 - 201; http://dx.doi.org/10.1091/mbc.E10-03-0256; PMID: 21119010
- Ramalingam N, Zhao H, Breitsprecher D, Lappalainen P, Faix J, Schleicher M. Phospholipids regulate localization and activity of mDia1 formin. Eur J Cell Biol 2010; 89:723 - 32; http://dx.doi.org/10.1016/j.ejcb.2010.06.001; PMID: 20619927
- Alberts AS. Identification of a carboxyl-terminal diaphanous-related formin homology protein autoregulatory domain. J Biol Chem 2001; 276:2824 - 30; http://dx.doi.org/10.1074/jbc.M006205200; PMID: 11035012
- Williams CL. The polybasic region of Ras and Rho family small GTPases: a regulator of protein interactions and membrane association and a site of nuclear localization signal sequences. Cell Signal 2003; 15:1071 - 80; http://dx.doi.org/10.1016/S0898-6568(03)00098-6; PMID: 14575862
- Dubash AD, Guilluy C, Srougi MC, Boulter E, Burridge K, García-Mata R. The small GTPase RhoA localizes to the nucleus and is activated by Net1 and DNA damage signals. PLoS One 2011; 6:e17380; http://dx.doi.org/10.1371/journal.pone.0017380; PMID: 21390328
- Michaelson D, Abidi W, Guardavaccaro D, Zhou M, Ahearn I, Pagano M, Philips MR. Rac1 accumulates in the nucleus during the G2 phase of the cell cycle and promotes cell division. J Cell Biol 2008; 181:485 - 96; http://dx.doi.org/10.1083/jcb.200801047; PMID: 18443222
- Kovar DR, Harris ES, Mahaffy R, Higgs HN, Pollard TD. Control of the assembly of ATP- and ADP-actin by formins and profilin. Cell 2006; 124:423 - 35; http://dx.doi.org/10.1016/j.cell.2005.11.038; PMID: 16439214
- Mellad JA, Warren DT, Shanahan CM. Nesprins LINC the nucleus and cytoskeleton. Curr Opin Cell Biol 2011; 23:47 - 54; http://dx.doi.org/10.1016/j.ceb.2010.11.006; PMID: 21177090
- Holaska JM, Kowalski AK, Wilson KL. Emerin caps the pointed end of actin filaments: evidence for an actin cortical network at the nuclear inner membrane. PLoS Biol 2004; 2:E231; http://dx.doi.org/10.1371/journal.pbio.0020231; PMID: 15328537
- Simon DN, Zastrow MS, Wilson KL. Direct actin binding to A- and B-type lamin tails and actin filament bundling by the lamin A tail. Nucleus 2010; 1:264 - 72; http://dx.doi.org/10.4161/nucl.1.3.11799; PMID: 21327074
- Jégou A, Carlier M-F, Romet-Lemonne G. Formin mDia1 senses and generates mechanical forces on actin filaments. Nat Commun 2013; 4:1883; http://dx.doi.org/10.1038/ncomms2888; PMID: 23695677
- Courtemanche N, Lee JY, Pollard TD, Greene EC. Tension modulates actin filament polymerization mediated by formin and profilin. Proc Natl Acad Sci U S A 2013; 110:9752 - 7; http://dx.doi.org/10.1073/pnas.1308257110; PMID: 23716666
- Vartiainen MK, Guettler S, Larijani B, Treisman R. Nuclear actin regulates dynamic subcellular localization and activity of the SRF cofactor MAL. Science 2007; 316:1749 - 52; http://dx.doi.org/10.1126/science.1141084; PMID: 17588931
- Mouilleron S, Langer CA, Guettler S, McDonald NQ, Treisman R. Structure of a pentavalent G-actin*MRTF-A complex reveals how G-actin controls nucleocytoplasmic shuttling of a transcriptional coactivator. Sci Signal 2011; 4:ra40; http://dx.doi.org/10.1126/scisignal.2001750; PMID: 21673315
- Pawłowski R, Rajakylä EK, Vartiainen MK, Treisman R. An actin-regulated importin α/β-dependent extended bipartite NLS directs nuclear import of MRTF-A. EMBO J 2010; 29:3448 - 58; http://dx.doi.org/10.1038/emboj.2010.216; PMID: 20818336
