Abstract
The DNA assisted solid-phase proximity ligation assay (SP-PLA) provides a unique opportunity to specifically detect prion protein (PrP) aggregates by investigating the collocation of 3 or more copies of the specific protein. We have developed an SP-PLA that can detect PrP aggregates in brain homogenates from infected hamsters even after a 107-fold dilution. In contrast, brain homogenate from uninfected animals did not generate a detectable signal at 100-fold higher concentration. Using either of the 2 monoclonal anti-PrP antibodies, 3F4 and 6H4, we successfully detected low concentrations of aggregated PrP. The presented results provide a proof of concept that this method might be an interesting tool in the development of diagnostic approaches of prion diseases.
Abbreviations
| BSE | = | bovine spongiform encephalopathy |
| CJD | = | Creutzfeldt-Jakob disease |
| CSF | = | cerebrospinal fluid |
| FIDA | = | fluorescence intensity distribution analysis |
| qPCR | = | quantitative real-time PCR |
| QuIC | = | quaking-induced conversion |
| PLA | = | proximity ligation assay |
| PMCA | = | protein misfolding cyclic amplification |
| PrP | = | prion protein |
| PrPC | = | cellular prion protein |
| PrPSc | = | scrapie prion protein |
| SP-PLA | = | solid phase proximity ligation assay |
Prion diseases are fatal infectious neurodegenerative diseases of humans and animals.Citation1 During the asymptomatic phase, which can last for decades in humans, the disease can be transmitted between animals and humans. There is a critical need for early diagnostics in order to take measures to prevent the spread of prion diseases. The key event in prion disease pathogenesis is the conformational conversion of the normal cellular prion protein (PrPC) into an aggregated multimeric, protease resistant form called the scrapie prion protein (PrPSc). There are several methods to distinguish and separate PrPSc from PrPC. Classical methods to detect PrPSc require digestion with proteinase K, followed by immunoassays such as western blot or ELISA. A PrPSc detection assay with increased sensitivity has been reported using immuno-PCR,Citation2,3 where bound antibodies are detected by amplification of conjugated DNA oligonucleotides. However, due to difficulties of raising high quality antibodies that can distinguish PrPSc from PrPC, proteinase K treatment is required to ensure specificity of the assay. Also, proteinase K-sensitive disease-related species of PrP have been foundCitation4,5 and it is therefore important to develop new sensitive and specific methods to detect misfolded PrP without the need for treatment with proteinase K. The fact that infectious PrP can propagate itself has been the basis for development of methods that demonstrate the presence of aggregating protein via in vitro amplification of the misfolded protein. Protein misfolding cyclic amplification (PMCA)Citation6 is a method where the accumulation of aggregated PrPSc from monomers is triggered by the presence of PrPSc in a sample, serving to seed the conversion of PrPC from brain extracts into large amounts of PrPSc. Several assays have been developed based on PMCA for detection of PrPSc in blood,Citation7-10 but the assays are not easily adapted to clinical settings due to the slow and complex procedure, high levels of false positives, and potential health hazards. The quaking-induced conversion (QuIC) assay similarly uses recombinant PrPC that is induced by PrPSc to convert into amyloid fibrils, but in a shorter amount of time compared to PMCA.Citation11,12 QuIC has been used for detection of PrPSc in human cerebrospinal fluid (CSF)Citation13 to distinguish patients with Creutzfeldt-Jakob disease (CJD) from healthy controls or patients with other neurodegenerative diseases with 100% specificity and a sensitivity around 80%. The sensitivity of the assay has been further enhanced by improving the QuIC protocol and adding an immunoprecipitation step with the PrPSc selective antibody 15B3Citation14,15 prior to the assay.Citation16 Other ways to specifically capture and separate PrPSc from other sample components and from PrPC include binding to a polymeric compound (Seprion ligands),Citation17 a method that has been the basis for the development of immunoassays sufficiently sensitive to detect PrPSc in whole blood from humans with variant CJD.Citation18 Yet another method to separate PrPSc from PrPC is through precipitation with sodium phosphotungstate (NaPTA).Citation19 This procedure has been combined with a technique based on fluorescence intensity distribution analysis (surface-FIDA) to detect PrPSc in CSF from cattle with bovine spongiform encephalopathy (BSE)Citation20 and blood plasma from scrapie-infected sheep.Citation21 The FIDA assay detects PrPSc based on the fact that they are aggregates of large numbers of copies of a protein. A fluorescence labeled monoclonal antibody is allowed to bind the target and only when many antibodies are bound in close proximity, i.e. to the same aggregate, will this generate a detectable signal:Citation22 A monoclonal antibody can only bind once per monomeric PrPC, but several antibodies can bind to aggregates of PrPSc, leading to a concentration of fluorophores on PrPSc that can be detected using fluorescence correlation spectroscopy over the randomly distributed antibodies in solution or bound to PrPC.
Herein we describe a sensitive method to detect aggregated PrP, which is based on the principle that amplifiable reporter DNA molecules are only generated when 3 copies of a monoclonal antibody bind 3 or more identical epitopes in proximity, such as by binding an aggregate of a target protein. The specific detection of aggregated PrP is based on the solid-phase proximity ligation assay (SP-PLA).Citation23,24 In SP-PLA targeted proteins are first captured on a solid support via immobilized antibodies before addition of 2 PLA probes, that is antibodies with conjugated oligonucleotides, followed by washes, ligation of oligonucleotides on pairs of antibodies that have bound in proximity, and amplified detection by quantitative real-time PCR (qPCR). If all 3 affinity reagents required for detection are directed against the same epitope, then the assay can be used to detect aggregated forms of a protein recognized by the antibody. We have previously demonstrated the utility of this assay mechanism to detect Aβ-oligomers or protofibrils, thought to herald the onset of Alzheimer disease, by using a single Aβ-protofibril-specific monoclonal antibody for all 3 binding events in SP-PLA.Citation25
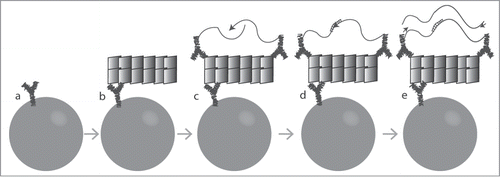
We established a SP-PLA protocol for detection of aggregated PrP using either monoclonal antibodies 3F4Citation26 or 6H4,Citation14 both well known to recognize the PrP protein (). Briefly, a monoclonal antibody was immobilized on magnetic beads and used for enrichment of PrP from biological samples. The same monoclonal antibody was also coupled to 2 different DNA oligonucleotides in separate reactions and this pair was allowed to bind captured proteins. Only aggregates of 3 or more PrP subunits can host 2 DNA-coupled antibodies with different, ligatable sequences after capture, as required to generate a signal in the assay.
Figure 1. Schematic illustration of SP-PLA. (A) Captured antibodies are immobilized on paramagnetic beads. (B) When the sample is incubated with the beads, the targeted PrPs are captured. (C) The same clone of antibody is combined with streptavidin-DNA oligonucleotide conjugates to create PLA probes that are incubated with the captured targets. (D) Oligonucleotides attached to antibodies that have bound in close proximity to each other are joined by enzymatic ligation in the presence of a connector DNA oligonucleotide. (E) Only ligated oligonucleotide reporter molecules can be amplified, detected, and quantified by real-time quantitative PCR.
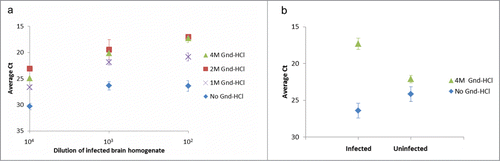
It has previously been shown that PrPC and PrPSc exhibit different accessible epitopes according to their respective confirmations, and that treatment with denaturing agents such as guanidine-HCl increases the reactivity of certain PrPSc epitopes contrarily to PrPC.Citation19,27,28 This phenomenon was observed with our SP-PLA protocol, increasing the sensitivity of this technique (). As the SP-PLA requires triple recognition events our observations suggest that denaturation with high concentrations of guanidine-HCl would induce incomplete disaggregation of PrPSc as previously described,Citation29-31 with maintenance of at least trimers or small aggregates. No significant change in signal was detected upon denaturation of brain homogenate from uninfected hamster ().
Figure 2. Effect of denaturation of hamster brain homogenates with guanidine-HCl (Gnd-HCl) prior to SP-PLA. (A) A homogenate from an infected hamster brain was incubated with different concentrations of Gnd-HCl and with a control without Gnd-HCl. Dilution series of the homogenates were analyzed for PrPSc through SP-PLA using the monoclonal antibody 3F4. (B) The effect of Gnd-HCl was also examined for a 100-fold dilution of infected and uninfected brain homogenate. Plotted are the mean cycle numbers at threshold (Ct) with standard deviations from duplicate experiments.
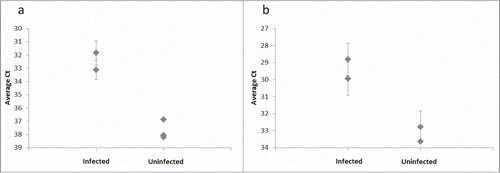
As a proof-of-concept experiment we used the assay to compare brain homogenates from hamsters infected with the prion strain 263K and from uninfected control animals. Using the 3F4 antibody, we could successfully distinguish between brain homogenate from hamster infected with strain 263K and brain homogenate from uninfected hamster, as the infected sample gave detection signals at 100-fold lower concentration than the sample obtained from uninfected hamster (). To extend our results we assayed the same brain homogenates in parallel with brain homogenate from a different infected hamster and from 2 more uninfected hamsters. The results show that the assay clearly distinguished both infected samples from all 3 uninfected controls at a 10 million-fold dilution (). Using the antibody 6H4 we could similarly distinguish infected lysates from those from uninfected controls when both were assayed at a 1 million-fold dilution ().
Figure 3. Distinguishing PrPSc aggregates in brain homogenate from an infected hamster vs. an uninfected control hamster. Dilutions of homogenates from brains of infected or uninfected hamsters in buffer were analyzed for the presence of PrPSc using SP-PLA with the 3F4 monoclonal antibody. A cut-off value was set at 3 standard deviations above the mean signal from a negative control (only buffer). Plotted in the graph are the ratios of reporter DNA molecules in the assayed samples relative to this cut off. The data shown are the mean values of 3 replicates with standard deviations.
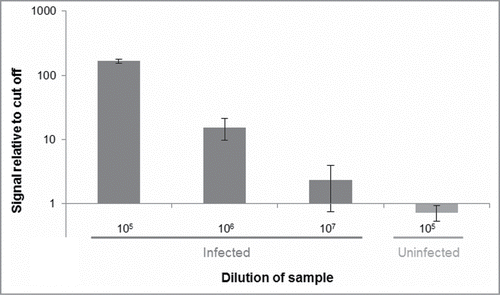
Figure 4. Distinguishing PrPSc aggregates in brain homogenate from several infected and uninfected hamsters. (A) The 3F4 SP-PLA was used to detect PrPSc aggregates in brain homogenates from 2 infected hamsters and from 3 uninfected hamsters diluted 10 million times. (B) An SP-PLA based on the monoclonal antibody 6H4 was used to detect PrPSc aggregates in the same samples diluted 1 million times. Each sample was assayed in duplicates. Plotted are the mean cycle numbers at threshold (Ct) with standard deviations.
The results prove the validity of the concept of using SP-PLA for sensitive detection of aggregated PrP by requiring several antibody-epitope binding events per target aggregate. Here, we successfully developed assays using either the monoclonal antibodies 3F4 or 6H4, suggesting that the assays could also be adapted for detection of aggregated PrP from other species using the appropriate monoclonal antibodies. The SP-PLA also provides a novel method to study the actual aggregation state of different prion strains treated with different denaturants or other possible decontaminants.
For each reaction we only use 5 μl of sample. The capture to a solid support also allows the assay to be adapted for analyses of larger sample volumes in order to further increase the sensitivity, as has previously been used to improve the sensitivity of both immunoassaysCitation18,32 and QuICCitation16 for detection of PrPSc, and PLA-based detection of Aβ protofibrils.Citation25 We have previously shown that SP-PLA shows similar assay performance in whole blood as in buffer or plasma.Citation24 The only instrumentation required for the assay is a qPCR machine, which represents standard laboratory instrumentation. We therefore believe that the assay given further optimization will be suitable for routine analyses of samples from body fluids such as CSF or blood, allowing ante mortem testing.
Materials and Methods
Reagents
The monoclonal antibody 3F4Citation26 was biotinylated using the ChromaLink™ biotin labeling kit (B-9007-105K, Solulink). The reaction was performed with 100 μl 3F4 antibody at a concentration of 0.9 mg/mL, and a 20-fold molar excess of ChromaLink biotin over the antibodies. The biotinylated monoclonal antibody 6H4 (01-043) was purchased from Prionics.
The oligonucleotidestreptavidin conjugates SLC1 (streptavidin5′CGCATCGCCCTTGGACTACGACTGACGAACCGCTTTGCCTGACTGATCGCTAAATCGTG3′) and SLC2 (5′TCGTGTCTAAAGTCCGTTACCTTGATTCCCCTAACCCTCTTGAAAAATTCGGCATCGGTGA-3′-streptavidin) were custom ordered from Solulink. Prior to use the streptavidin-oligonucleotide conjugates were treated with a 3-fold molar excess of free streptavidin (S4762, Sigma-Aldrich) at 65°C for 30 min as previously described.Citation24,33
The connector oligonucleotide (5′-TACTTAGACACGACACGATTTAGTTT-3′) serving as template for the ligation, the forward (5′CATCGCCCTTGGACTACGA-3′) and reverse (5′-GGGAATCAAGGTAACGGACTTTAG-3′) primers were all purchased from Integrated DNA Technology. The TaqMan probe (FAM-5′TGACGAACCGCTTTGCCTGA-3′-MGB[quencher]) was purchased from Applied Biosystems.
Preparation of hamster samples
Brains from 263K-infected hamsters were sampled at the terminal stage of the disease. Brains from the infected and uninfected animals were homogenized at 20% (weight/volume) in a 5% glucose solution.
Denaturing the samples
The brain homogenate samples were denatured by mixing the sample with guanidine-HCl solution 8 M in H2O (G9284, Sigma-Aldrich) at a 1:1 ratio. When titrating the concentration of guanidine-HCl the samples were mixed 1:1 with 8M, 4M, and 2M guanidine-HCl in H2O respectively. The samples were incubated at room temperature (RT) for ten minutes before they were further diluted at least 10-fold in PLA buffer (1 mM D-biotin (B-1595, Life Technologies), 0.1% purified bovine serum albumin (A7888, Sigma-Aldrich), 0.05% Tween 20 (P9416, Sigma-Aldrich), 100 nM goat IgG (I5256, Sigma-Aldrich), 0.1 μg/μl salmon sperm DNA (15632-011, Life Technologies), 5 mM EDTA (E5134, Sigma-Aldrich), 1xPBS).
Solid-phase proximity ligation assay
SP-PLA was performed as previously described.Citation24,34 Briefly, 1.5 μg of the biotinylated antibody – either 3F4 or 6H4 – was immobilized on 1 mg streptavidin-coated magnetic beads (Dynabeads MyOne Streptavidin T1; 65602, Life Technologies). Five μl of denatured brain homogenate sample was diluted ten times in PLA buffer and thereafter mixed with the beads to allow capture of the target molecules. After 1.5 h incubation under rotation at RT the beads were separated from the supernatant using a DynaMag-96 Side magnet (65602, Life Technologies). After washing once with 100 μl 0.02% N-lauroylsarcosine sodium salt solution (61747, Sigma-Aldrich) in 1xPBS, and twice with 100 μl washing buffer (1xPBS, 0.05% Tween 20), 250 pM of the pair of PLA probes in PLA buffer were added. These probes consisted in the same monoclonal antibody as that used for capture mixed at a 1:1 ratio with 2 streptavidin-oligonucleotide conjugates, one having oligonucleotides with free phosphorylated 5’ ends and one with free 3’ ends. The probes were allowed to find their targets during 1.5 h incubation at RT under rotation. The beads were washed twice in 100 μl washing buffer before adding 50 μl ligation and PCR mix (1xPCR buffer, 2.5 mM MgCl2 , 1.5 units Platinum Taq Polymerase (10966, Life Technologies), 0.1 μM of each primer, 0.2 μM TaqMan probe, 0.08 mM ATP (R0441, Thermo Scientific), 0.1 μM connector oligonucleotide, 0.2 mM dNTPs (with dTTP substituted by dUTP; V0195, Thermo Scientific), 0.5 units T4 DNA ligase (EL0016, Thermo Scientific), 0.1 units uracil-DNA glycocylase (UDG; EN0362, Thermo Scientific)). The ligation was carried out for 10 min at RT before starting the qPCR on an Mx3000 real-time PCR instrument (Stratagene), with an initial hot start at 95°C for 10 min, followed by 40 cycles of 15 s at 95°C and 60 s at 60°C.
Data analysis
The qPCR data was analyzed with the MxPro software (Stratagene) to extract the cycle of threshold (Ct) values. To set a cut off value for the assay we used the Ct value of a negative control sample (only PLA buffer) and subtracted 3 standard deviations from the average Ct value for triplicate assay. Based on this cut off, we calculated the ratio of ligated DNA reporter molecules in the samples relative to the amount of reporter molecules set as cut off for the assay.
Signal relative to cut off = 2Ct(cut off)-Ct(sample)
Disclosure of Potential Conflicts of Interest
UL is a cofounder and shareholder of Olink Bioscience that commercializes the proximity ligation assay technology described in this paper.
Funding
This work was funded by the Knut and Alice Wallenberg Foundation, FORMAS (2006-2856, MKM), Alliance Biosecure Foundation, Åke Wiberg Foundation, IngaBritt and Arne Lundberg's Foundation, European Science Foundation, the Swedish Research Council for medicine (2007-2720, UL), and by the European Community's 6th and 7th Framework Programs.
References
- Prusiner SB. Novel proteinaceous infectious particles cause scrapie. Science 1982; 216:136-44; PMID:6801762; http://dx.doi.org/10.1126/science.6801762
- Gofflot S, Deprez M, Moualij B el, Osman A, Thonnart J-F, Hougrand O, Heinen E, Zorzi W. Immunoquantitative PCR for prion protein detection in sporadic creutzfeldt–jakob disease. Clin Chem 2005; 51:1605-11; PMID:16002456; http://dx.doi.org/10.1373/clinchem.2005.050120
- Barletta JM, Edelman DC, Highsmith WE, Constantine NT. Detection of ultra-low levels of pathologic prion protein in scrapie infected hamster brain homogenates using real-time immuno-PCR. J Virol Methods 2005; 127:154-64; PMID:15921765; http://dx.doi.org/10.1016/j.jviromet.2005.04.007
- Bessen RA, Marsh RF. Biochemical and physical properties of the prion protein from two strains of the transmissible mink encephalopathy agent. J Virol 1992; 66:2096-101; PMID:1347795
- Tzaban S, Friedlander G, Schonberger O, Horonchik L, Yedidia Y, Shaked G, Gabizon R, Taraboulos A. Protease-sensitive scrapie prion protein in aggregates of heterogeneous sizes. Biochemistry 2002; 41:12868-75; PMID:12379130; http://dx.doi.org/10.1021/bi025958g
- Saborio GP, Permanne B, Soto C. Sensitive detection of pathological prion protein by cyclic amplification of protein misfolding. Nature 2001; 411:810-3; PMID:11459061; http://dx.doi.org/10.1038/35081095
- Castilla J, Saá P, Soto C. Detection of prions in blood. Nat Med 2005; 11:982-5; PMID:16127436
- Saá P, Castilla J, Soto C. Presymptomatic Detection of Prions in Blood. Science 2006; 313:92-4; http://dx.doi.org/10.1126/science.1129051
- Rubenstein R, Chang B, Gray P, Piltch M, Bulgin MS, Sorensen-Melson S, Miller MW. A novel method for preclinical detection of PrPSc in blood. J Gen Virol 2010; 91:1883-92; PMID:20357038; http://dx.doi.org/10.1099/vir.0.020164-0
- Trieschmann L, Santos AN, Kaschig K, Torkler S, Maas E, Schätzl H, Böhm G. Ultra-sensitive detection of prion protein fibrils by flow cytometry in blood from cattle affected with bovine spongiform encephalopathy. BMC Biotechnol 2005; 5:26; PMID:16202155; http://dx.doi.org/10.1186/1472-6750-5-26
- Atarashi R, Wilham JM, Christensen L, Hughson AG, Moore RA, Johnson LM, Onwubiko HA, Priola SA, Caughey B. Simplified ultrasensitive prion detection by recombinant PrP conversion with shaking. Nat Meth 2008; 5:211-2; http://dx.doi.org/10.1038/nmeth0308-211
- Orrú CD, Wilham JM, Hughson AG, Raymond LD, McNally KL, Bossers A, Ligios C, Caughey B. Human variant Creutzfeldt-Jakob disease and sheep scrapie PrP(res) detection using seeded conversion of recombinant prion protein. Protein Eng Des Sel 2009; 22:515-21; PMID:19570812; http://dx.doi.org/10.1093/protein/gzp031
- Atarashi R, Satoh K, Sano K, Fuse T, Yamaguchi N, Ishibashi D, Matsubara T, Nakagaki T, Yamanaka H, Shirabe S, et al. Ultrasensitive human prion detection in cerebrospinal fluid by real-time quaking-induced conversion. Nat Med 2011; 17:175-8; PMID:21278748; http://dx.doi.org/10.1038/nm.2294
- Korth C, Stierli B, Streit P, Moser M, Schaller O, Fischer R, Schulz-Schaeffer W, Kretzschmar H, Raeber A, Braun U, et al. Prion (PrPSc)-specific epitope defined by a monoclonal antibody. Nature 1997; 390:74-7; PMID:9363892; http://dx.doi.org/10.1038/36337
- Biasini E, Tapella L, Mantovani S, Stravalaci M, Gobbi M, Harris DA, Chiesa R. Immunopurification of Pathological Prion Protein Aggregates. PLoS ONE 2009; 4:e7816; PMID:19915706; http://dx.doi.org/10.1371/journal.pone.0007816
- Orrú CD, Wilham JM, Raymond LD, Kuhn F, Schroeder B, Raeber AJ, Caughey B. Prion disease blood test using immunoprecipitation and improved quaking-induced conversion. MBio 2011; 2:e00078-00011; PMID:21558432
- Lane A, Stanley CJ, Dealler S, Wilson SM. Polymeric Ligands with Specificity for Aggregated Prion Proteins. Clin Chem 2003; 49:1774-5; http://dx.doi.org/10.1373/49.10.1774
- Edgeworth JA, Farmer M, Sicilia A, Tavares P, Beck J, Campbell T, Lowe J, Mead S, Rudge P, Collinge J, et al. Detection of prion infection in variant Creutzfeldt-Jakob disease: a blood-based assay. The Lancet 2011; 377:487-93; http://dx.doi.org/10.1016/S0140-6736(10)62308-2
- Safar J, Wille H, Itri V, Groth D, Serban H, Torchia M, Cohen FE, Prusiner SB. Eight prion strains have PrP(Sc) molecules with different conformations. Nat Med 1998; 4:1157-65; PMID:9771749; http://dx.doi.org/10.1038/2654
- Birkmann E, Henke F, Weinmann N, Dumpitak C, Groschup M, Funke A, Willbold D, Riesner D. Counting of single prion particles bound to a capture-antibody surface (surface-FIDA). Vet Microbiol 2007; 123:294-304; PMID:17499942; http://dx.doi.org/10.1016/j.vetmic.2007.04.001
- Bannach O, Birkmann E, Reinartz E, Jaeger K-E, Langeveld JPM, Rohwer RG, Gregori L, Terry LA, Willbold D, Riesner D. Detection of prion protein particles in blood plasma of scrapie infected sheep. PLoS ONE 2012; 7:e36620; PMID:22567169; http://dx.doi.org/10.1371/journal.pone.0036620
- Pitschke M, Prior R, Haupt M, Riesner D. Detection of single amyloid β-protein aggregates in the cerebrospinal fluid of Alzheimer's patients by fluorescence correlation spectroscopy. Nat Med 1998; 4:832-4; PMID:9662376; http://dx.doi.org/10.1038/nm0798-832
- Fredriksson S, Gullberg M, Jarvius J, Olsson C, Pietras K, Gústafsdóttir SM, Ostman A, Landegren U. Protein detection using proximity-dependent DNA ligation assays. Nat Biotechnol 2002; 20:473-7; PMID:11981560; http://dx.doi.org/10.1038/nbt0502-473
- Darmanis S, Nong RY, Hammond M, Gu J, Alderborn A, Vänelid J, Siegbahn A, Gustafsdottir S, Ericsson O, Landegren U, et al. Sensitive plasma protein analysis by microparticle-based proximity ligation assays. Mol Cell Proteomics 2010; 9:327-35; PMID:19955079; http://dx.doi.org/10.1074/mcp.M900248-MCP200
- Kamali-Moghaddam M, Pettersson FE, Wu D, Englund H, Darmanis S, Lord A, Tavoosidana G, Sehlin D, Gustafsdottir S, Nilsson LNG, et al. Sensitive detection of Aβ protofibrils by proximity ligation-relevance for Alzheimer's disease. BMC Neurosci 2010; 11:124; PMID:20923550; http://dx.doi.org/10.1186/1471-2202-11-124
- Kascsak RJ, Rubenstein R, Merz PA, Tonna-DeMasi M, Fersko R, Carp RI, Wisniewski HM, Diringer H. Mouse polyclonal and monoclonal antibody to scrapie-associated fibril proteins. J Virol 1987; 61:3688-93; PMID:2446004
- Bibby DF, Gill AC, Kirby L, Farquhar CF, Bruce ME, Garson JA. Application of a novel in vitro selection technique to isolate and characterise high affinity DNA aptamers binding mammalian prion proteins. J Virol Methods 2008; 151:107-15; PMID:18433888; http://dx.doi.org/10.1016/j.jviromet.2008.03.013
- Xiao SJ, Hu PP, Wu XD, Zou YL, Chen LQ, Peng L, Ling J, Zhen SJ, Zhan L, Li YF, et al. Sensitive discrimination and detection of prion disease-associated isoform with a dual-aptamer strategy by developing a sandwich structure of magnetic microparticles and quantum dots. Anal Chem 2010; 82:9736-42; PMID:21038863; http://dx.doi.org/10.1021/ac101865s
- Sokolowski F, Modler AJ, Masuch R, Zirwer D, Baier M, Lutsch G, Moss DA, Gast K, Naumann D. Formation of critical oligomers is a key event during conformational transition of recombinant syrian hamster prion protein. J Biol Chem 2003; 278:40481-92; PMID:12917432; http://dx.doi.org/10.1074/jbc.M304391200
- Novitskaya V, Makarava N, Bellon A, Bocharova OV, Bronstein IB, Williamson RA, Baskakov IV. Probing the conformation of the prion protein within a single amyloid fibril using a novel immunoconformational assay. J Biol Chem 2006; 281:15536-45; PMID:16569635; http://dx.doi.org/10.1074/jbc.M601349200
- Dong J, Castro CE, Boyce MC, Lang MJ, Lindquist S. Optical trapping with high forces reveals unexpected behaviors of prion fibrils. Nat Struct Mol Biol 2010; 17:1422-30; PMID:21113168; http://dx.doi.org/10.1038/nsmb.1954
- Tattum MH, Jones S, Pal S, Khalili-Shirazi A, Collinge J, Jackson GS. A highly sensitive immunoassay for the detection of prion-infected material in whole human blood without the use of proteinase K. Transfusion 2010; 50:2619-27; PMID:20561299; http://dx.doi.org/10.1111/j.1537-2995.2010.02731.x
- Darmanis S, Kähler A, Spångberg L, Kamali-Moghaddam M, Landegren U, Schallmeiner E. Self-assembly of proximity probes for flexible and modular proximity ligation assays. BioTechniques 2007; 43:443-444, 446, 448 passim; PMID:18019334; http://dx.doi.org/10.2144/000112551
- Nong RY, Wu D, Yan J, Hammond M, Gu GJ, Kamali-Moghaddam M, Landegren U, Darmanis S. Solid-phase proximity ligation assays for individual or parallel protein analyses with readout via real-time PCR or sequencing. Nat Protocols 2013; 8:1234-48; http://dx.doi.org/10.1038/nprot.2013.070
