Abstract
The generation of intracellular microbe-associated molecular pattern (MAMP)-triggered Ca2+ transients was recently demonstrated to involve ionotropic Glutamate Receptor (iGluR)-like channels in Arabidopsis and tobacco. Here we elaborate on our previous findings and refine our insights in the putative agonist binding profile and potential mode of desensitization of MAMP-activated plant iGluRs. Based on results from pharmacological inhibition and desensitization experiments, we propose that plant iGluR complexes responsible for the MAMP-triggered Ca2+ signature have a binding profile that combines the specificities of mammalian NMDA-and non-NMDA types of iGluRs, possibly reflecting the evolutionary history of plant and animal iGluRs. We further hypothesize that, analogous to the mammalian NMDA-NR1 receptor, desensitization of plant iGluR-like channels might involve binding of the ubiquitous Ca2+ sensor calmodulin to a cytoplasmic C-terminal domain.
iGluR-Like Channel Function in Plant Defense and Development
Ionotropic Glutamate Receptor (iGluR)-like channels represent a family of integral membrane proteins that control amino acid-gated cation fluxes. This protein class is best known for its key role in vertebrate neurotransmission.Citation1 Recently, two groups demonstrated the involvement of iGluR-like channels in microbe-associated molecular pattern (MAMP)-triggered Ca2+ fluxes in plants. In the presence of iGluR channel agonists and antagonists, the MAMP-induced Ca2+ transient was considerably reduced and downstream responses were attenuated both in tobacco (Nicotiana tabacum) cell cultures in response to cryptogeinCitation2 and intact Arabidopsis thaliana seedlings in response to flg22, elf18 and chitin, respectively.Citation3 These altered responses include mitogen-activated protein kinase (MAPK) activation, transcriptional regulation of downstream response genesCitation3 and nitric oxide production.Citation2 In addition, cryptogein induces exocytosis-dependent accumulation of glutamate in the apoplastic space.Citation2 Being present as a family with 20 members in Arabidopsis,Citation4 the acquisition of genetic evidence corroborating the role of these channels in MAMP-triggered Ca2+ influx is cumbersome and will require extensive future work. These receptors likely function as heterotypic quaternary complexes, therefore the members might compensate for each other’s function in the complex. Results from desensitization experiments, using several amino acids in glr3.3 and glr3.4 single or double mutants, suggested that multiple subunits together control amino acid-dependent Ca2+ influx. These data fit a model where various quaternary complexes of iGluR-like channels with dissimilar sensitivity to different amino acids co-exist.Citation5
Mutants in some iGluR family members, either as single knockout or mutant combinations, show altered Ca2+ responses or developmental phenotypes. For example, glr3.3 mutants exhibit more variable root growth compared with wild type plants upon a gravity stimulusCitation6 and glr1.2 and glr3.7 mutants have altered Ca2+ responses in pollen tubes and altered pollen tube morphology and growth speed, respectively.Citation7 Overexpression of AtGLUR2/GLR3.1 alters Ca2+ homeostasis and utilization and renders plants sensitive to ion stress.Citation8 Moreover, GLR1.1 is involved in stomatal closure and abscisic acid signaling and contributes to fungal disease resistance.Citation9,Citation10
Ligand Preference of Arabidopsis iGluRs Contributing to MAMP-Triggered Ca2+ Transients
In mammalian systems, three main subclasses of iGluR-like channels with ion gating activity are distinguished, partly based on their selective preference for synthetic/non-native agonists: the NMDA (N-Methyl-d-aspartic acid), the AMPA (2-amino-3-(5- methyl-3-oxo-1,2-oxazol-4-yl)propanoic acid) and the kainate-binding receptor channels. The AMPA and kainate receptor channels are generally grouped together as the non-NMDA class.Citation11-Citation13 In the native situation, mammalian NMDA receptor channels are activated by L-aspartate and L-glutamate and additionally require binding of the co-agonists d-serine or glycine at an allosteric binding site for activity.Citation1,Citation11,Citation13-Citation15
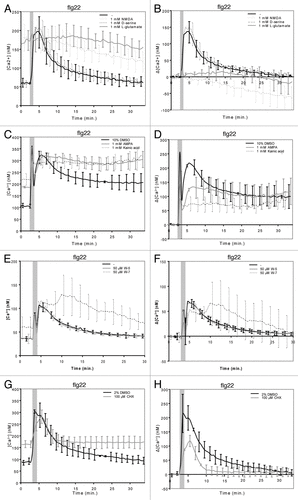
We studied if the plant repertoire of iGluR channels found to be involved in MAMP-triggered Ca2+ influx shows any preference toward these synthetic agonists using the desensitization assays recently described by Kwaaitaal and coworkers.Citation3 Pre-treatment of transgenic Arabidopsis seedlings expressing the Ca2+ sensor aequorin with the agonists prior to MAMP addition revealed that NMDA, like L-glutamate and L-aspartate, strongly inhibits the flg22-induced Ca2+ response and increases steady-state Ca2+ levels (see ref. Citation3, Figure 1A and B). Using the phospholipase C inhibitor U-73122 as precedence, we previously showed that an increase in steady-state [Ca2+] does not per se inhibit a normal MAMP-triggered Ca2+ signature.Citation3 The non-NMDA agonists kainic acid and AMPA also reduced the flg22-induced Ca2+ transient and increased steady-state Ca2+ levels, but to a lesser extent than NMDA (). By contrast, pre-treatment with the NMDA receptor co-agonist d-serine did not alter Ca2+ responses to flg22 (). In sum, these results corroborate our and othersʼ previous findings indicating a role for iGluR-like channels in the generation of MAMP-induced Ca2+ transients.Citation2,Citation3 In addition, the data suggest that plant iGluR-like channel complexes have essentially a ligand binding profile that combines the specificities of both major classes of mammalian iGluR-like channels, the NMDA and the non-NMDA receptor channels. However, the effect of NMDA on the activity of Arabidopsis iGluRs seems to be more pronounced, suggesting a tendency toward a stronger responsiveness of the Arabidopsis iGluR complexes to NMDA-like agonists. This interpretation is further supported by previous data revealing that the NMDA receptor antagonists AP-5 and AP-7 cause a strong decrease in MAMP-induced Ca2+ transients, while the non-NMDA receptor antagonists CNQX and DNQX showed either no or a much weaker effect.Citation2,Citation3,Citation7 Our pharmacological classification of plant iGluRs according to their responsiveness to various inhibitors is consistent with the phylogenetic analysis of plant iGluR genes, in which plant iGluRs grouped separately from mammalian NMDA and non-NMDA type iGluRs and which indicated that the divergence of animal and plant iGluR genes likely predated the divergence of iGluR subtypes in animals.Citation4,Citation11
Figure 1. Ligand specificity of MAMP-stimulated iGluRs and molecular control of iGluR desensitization.10–12-d-old seedlings were pre-treated for 30 min with (co-) agonists, W-7 or CHX as described in Kwaaitaal et al. (2011) and then 1 μM flg22 was added to trigger a Ca2+ transient. Seedlings were pre-treated with (A, B) 1 mM NMDA, 1 mM d-serine, 1 mM L-glutamate, (C, D) 1 mM AMPA, 1 mM kainic acid, (E, F) 50 μM W-5 or W-7 or (G, H) 100 μM CHX. Shown are the absolute Ca2+ concentrations (A, C, E, G) or the background-corrected differences in cellular Ca2+ levels (B, D, F, H). Data given are the mean ± standard deviation of 6–12 seedlings per measurement and represent typical examples of three or more independent experiments. Note that the first short and strong Ca2+ peak marked by the gray bar results from the mechanical stimulation caused by the addition of the stimulus. Also note the large standard deviation of the W-7-treated seedlings compared with the control measurements, which reflects the high variation in the individual seedling curves upon treatment with this compound (see Fig. S2). For further experimental details see reference Citation3.
Control of iGluR Desensitization
After ligand binding and pore opening, iGluR-like channels are temporarily desensitized for a new response. This feature of individual iGluRs, together with Ca2+ efflux from the cytosol mediated by Ca2+-ATPases and anion exchange channels,Citation16 may contribute to shaping the integrated Ca2+ signature seen in experimental systems using the luminescent aequorin Ca2+ sensor. Several molecular modes of transient iGluR inactivation have been described, including structural rearrangements of the membrane-spanning and/or extracellular domains as well as binding of the Ca2+ sensor calmodulin (CaM) to the cytosolic C-terminus of the receptor channel.Citation12,Citation17-Citation20 Precedence for the latter is provided by the mammalian N-methyl-D-aspartate receptors (NMDARs), a class of iGluR-like channels that are major contributors to Ca2+ flux into brain neurons and that play a critical role in learning, memory, neural development and synaptic plasticity. NMDARs are composed of two families of subunits, termed NR1 and NR2. Upon Ca2+ influx, the C-terminal tail of NR1 subunits binds CaM to temporarily inactivate channel function and possibly to regulate receptor trafficking.Citation17,Citation20-Citation22
In the presence of Ca2+, CaM binding stimulates the formation of an amphipathic α-helix in otherwise largely unstructured regions of target proteins. The hydrophobic face of the helix then interacts with a hydrophobic pocket of Ca2+-CaM. For its interaction with the α-helix, Ca2+-CaM requires a series of properly spaced basic and hydrophobic residues for binding. From the analysis of a large number of CaM binding domains (CaMBDs), several classes of consensus sequences were deduced. Examples are the IQ class, the 1–8-14 and the 1–5-10 classes, where the numbers indicate the position of bulky hydrophobic residues that are needed for Ca2+-CaM binding.Citation23,Citation24
We examined the sequences of the 20 Arabidopsis iGluRs in silico for the presence of potential CaMBDs. In analogy to the location of the CaMBD in mammalian NMDA NR1 subunits, we focused our analysis on the C-terminal intracellular domain (Fig. Two and Fig. S1). We found that nine of the 20 Arabidopsis iGluRs possess a putative CaMBD in this region (). Notably, supposed CaMBDs were found in all iGluR subfamilies, suggesting that their presence might be a general feature of plant iGluR-like channels. However, for some iGluRs this region overlaps with the prediction of the most C-terminal transmembrane domain; in these instances the relevance of the CaMBD prediction is questionable. Absence of predicted CaMBDs in approximately half of the Arabidopsis iGluRs may either result from false-negative predictions or might indicate isoform-specific differences in the mode of receptor desensitization.
Figure 2. Presence of a potential CaMBD at the cytoplasmic C-terminus of Arabidopsis iGluRs. Transmembrane domain prediction was performed using Aramemnon (http://aramemnon.botanik.uni-koeln.de/) website and the stretches predicted using a hidden Markov modelCitation29 were marked. Putative CaMBDs (marked in gray) were identified based on the following web tool: http://calcium.uhnres.utoronto.ca/ctdb/ctdb/sequence.html.Citation23 For the prediction the last 268–375 amino acids were used, starting from the first TM domain and excluding the extracellular N-terminal domain. Marked residues represent those having a score higher than 6. Alignments were generated in the CLC Workbench package (www.clcbio.com).

Additional putative CaMBDs were predicted in the intracellular loop between the pore region (TM2) and TM3 and in the large extracellular domain between TM3 and TM4 (Fig. S1). The stretch of amino acids between TM2 and TM3 is relatively short and is localized between two membrane-embedded regions. This likely limits its mobility and probably hampers the formation of the amphipathic α-helix needed for Ca2+-CaM binding. Binding of CaM to extracellular binding sites has been reported and application of external CaM induces Ca2+ influx in plants.Citation25 Although potentially exciting, control of mammalian NMDARs by extracellular CaM binding has to our knowledge not been observed or studied, making the validity of these putative extracellular CaMBDs unlikely. In addition, the extracellular CaMBD predicted in GLR2.9 includes an amino acid (E) potentially involved in ligand binding.Citation14 This observation suggests that either also other features match the consensus sequences for CaMBDs or that the two domains overlap. Taken together, the predicted C-terminal regions remain the most likely CaMBD candidates in the Arabidopsis iGluR family.
To experimentally assess a potential role for CaM in controlling iGluR-mediated Ca2+ fluxes we pre-treated aequorin-expressing Arabidopsis seedlings with the well-characterized CaM binding inhibitor N-(6-aminohexyl)-5-chloro-1-naphthalenesulfonamide (W-7)Citation26 or its inactive analog N-(6-aminohexyl)-1-naphthalenesulfonamide (W-5).Citation27,Citation28 Upon W-7 application we found an increased steady-state [Ca2+] (), a slight inhibition of the Ca2+ transient in response to flg22 together with a broadening of the Ca2+ peak and an irregular recovery especially noticeable in individual seedling response curves (, Fig. S2A, B). By contrast, W-5-treated seedlings behaved like water-treated controls (, Fig. S2A, B). These observations suggest a direct or indirect CaM-dependent control of iGluR channel gating or closure. The broadening of the Ca2+ peak plus the irregular recovery could indicate that W-7 also interferes with CaM binding to other channel classes (e.g., cyclic nucleotide-gated channels) or affects Ca2+ efflux from the cytosol mediated by Ca2+-ATPases or anion exchange channels.Citation16
In a complementary experiment we used the protein biosynthesis inhibitor cycloheximide (CHX). We observed an increase in steady-state Ca2+ levels following such a pre-treatment that was similar in magnitude as the one observed upon treatment with W-7 or NMDA, but lower in magnitude than the one observed following treatment with L-glutamate, AMPA or kainate (; compare with ). In addition, the MAMP-induced Ca2+ transient was lower in magnitude, but had a similar shape as the curve of untreated seedlings (). These phenomena suggest that a class of proteins with a short half-life control the activity of Ca2+ in- or efflux channels in plants. Given the similarities in the effect of W-7 and CHX treatments on the flg22-triggered Ca2+ signature () one may speculate that inhibition of protein biosynthesis with CHX could affect cellular CaM levels. The increase in cytosolic [Ca2+] upon CHX treatment might thus reflect the loss of CaM-mediated control of Ca2+ channels and Ca2+ ATPases, thereby allowing more Ca2+ influx and/or preventing efficient Ca2+ efflux, respectively.
In conclusion, the prediction of CaMBDs in a subset of the Arabidopsis iGluR-like channels plus the altered MAMP-triggered Ca2+ responses in the presence of W-7 point toward a role CaM binding in plant iGluR control. To confirm that CaM binding indeed regulates iGluR activity, experiments testing the binding of CaM to iGluR-like channels are desired. The analysis of iGluR mutant variants that either lack the proposed CaMBD or harbour defective versions thereof promise to provide additional insights.
| Abbreviations: | ||
| AMPA | = | 2-amino-3-(5- methyl-3-oxo-1,2-oxazol-4-yl)propanoic acid |
| CaM | = | calmodulin |
| CaMBD | = | calmodulin binding domain |
| CHX | = | cycloheximide |
| iGluR | = | ionotropic glutamate receptor |
| MAMP | = | microbe-associated molecular pattern |
| NMDA | = | N-methyl-D-aspartic acid |
| W-5 | = | N-(6-aminohexyl)-1-aphthalenesulfonamide |
| W-7 | = | N-(6-aminohexyl)-5-chloro-1-naphthalenesulfonamide |
Additional material
Download Zip (502.8 KB)Acknowledgments
We acknowledge Tabea Marquardt for technical help in some of the experiments. The authors thank the Deutsche Forschungsgemeinschaft (DFG) Sonderforschungsbereich (SFB) 670, the Max-Planck Society and the Excellence Initiative of the German federal and state governments (seed fund provided by the RWTH Aachen University) for financial support.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- Paoletti P. Molecular basis of NMDA receptor functional diversity. Eur J Neurosci 2011; 33:1351 - 65; 10.1111/j.1460-9568.2011.07628.x; PMID: 21395862
- Vatsa P, Chiltz A, Bourque S, Wendehenne D, Garcia-Brugger A, Pugin A. Involvement of putative glutamate receptors in plant defence signaling and NO production. Biochimie 2011; 93:2095 - 101; 10.1016/j.biochi.2011.04.006; PMID: 21524679
- Kwaaitaal M, Huisman R, Maintz J, Reinstädler A, Panstruga R. Ionotropic glutamate receptor (iGluR)-like channels mediate MAMP-induced calcium influx in Arabidopsis thaliana.. Biochem J 2011; 440:355 - 65; 10.1042/BJ20111112; PMID: 21848515
- Chiu JC, Brenner ED, DeSalle R, Nitabach MN, Holmes TC, Coruzzi GM. Phylogenetic and expression analysis of the glutamate-receptor-like gene family in Arabidopsis thaliana.. Mol Biol Evol 2002; 19:1066 - 82; 10.1093/oxfordjournals.molbev.a004165; PMID: 12082126
- Stephens NR, Qi Z, Spalding EP. Glutamate receptor subtypes evidenced by differences in desensitization and dependence on the GLR3.3 and GLR3.4 genes. Plant Physiol 2008; 146:529 - 38; 10.1104/pp.107.108134; PMID: 18162597
- Miller ND, Durham Brooks TL, Assadi AH, Spalding EP. Detection of a gravitropism phenotype in glutamate receptor-like 3.3 mutants of Arabidopsis thaliana using machine vision and computation. Genetics 2010; 186:585 - 93; 10.1534/genetics.110.118711; PMID: 20647506
- Michard E, Lima PT, Borges F, Silva AC, Portes MT, Carvalho JE, et al. Glutamate receptor-like genes form Ca2+ channels in pollen tubes and are regulated by pistil D-serine. Science 2011; 332:434 - 7; 10.1126/science.1201101; PMID: 21415319
- Kim SA, Kwak JM, Jae SK, Wang MH, Nam HG. Overexpression of the AtGluR2 gene encoding an Arabidopsis homolog of mammalian glutamate receptors impairs calcium utilization and sensitivity to ionic stress in transgenic plants. Plant Cell Physiol 2001; 42:74 - 84; 10.1093/pcp/pce008; PMID: 11158446
- Humphry M, Bednarek P, Kemmerling B, Koh S, Stein M, Göbel U, et al. A regulon conserved in monocot and dicot plants defines a functional module in antifungal plant immunity. Proc Natl Acad Sci U S A 2010; 107:21896 - 901; 10.1073/pnas.1003619107; PMID: 21098265
- Kang S, Kim HB, Lee H, Choi JY, Heu S, Oh CJ, et al. Overexpression in Arabidopsis of a plasma membrane-targeting glutamate receptor from small radish increases glutamate-mediated Ca2+ influx and delays fungal infection. Mol Cells 2006; 21:418 - 27; PMID: 16819306
- Chiu J, DeSalle R, Lam HM, Meisel L, Coruzzi G. Molecular evolution of glutamate receptors: a primitive signaling mechanism that existed before plants and animals diverged. Mol Biol Evol 1999; 16:826 - 38; 10.1093/oxfordjournals.molbev.a026167; PMID: 10368960
- McFeeters RL, Oswald RE. Emerging structural explanations of ionotropic glutamate receptor function. FASEB J 2004; 18:428 - 38; 10.1096/fj.03-0873rev; PMID: 15003989
- Davenport R. Glutamate receptors in plants. Ann Bot 2002; 90:549 - 57; 10.1093/aob/mcf228; PMID: 12466095
- Dubos C, Huggins D, Grant GH, Knight MR, Campbell MM. A role for glycine in the gating of plant NMDA-like receptors. Plant J 2003; 35:800 - 10; 10.1046/j.1365-313X.2003.01849.x; PMID: 12969432
- Yang CR, Svensson KA. Allosteric modulation of NMDA receptor via elevation of brain glycine and D-serine: the therapeutic potentials for schizophrenia. Pharmacol Ther 2008; 120:317 - 32; 10.1016/j.pharmthera.2008.08.004; PMID: 18805436
- Bush DS. Calcium regulation in plant cells and its role in signaling. Annu Rev Plant Physiol Plant Mol Biol 1995; 46:95 - 122; 10.1146/annurev.pp.46.060195.000523
- Ataman ZA, Gakhar L, Sorensen BR, Hell JW, Shea MA. The NMDA receptor NR1 C1 region bound to calmodulin: structural insights into functional differences between homologous domains. Structure 2007; 15:1603 - 17; 10.1016/j.str.2007.10.012; PMID: 18073110
- Fay AM, Bowie D. Concanavalin-A reports agonist-induced conformational changes in the intact GluR6 kainate receptor. J Physiol 2006; 572:201 - 13; PMID: 16439423
- Wang C, Wang HG, Xie H, Pitt GS. Ca2+/CaM controls Ca2+-dependent inactivation of NMDA receptors by dimerizing the NR1 C termini. J Neurosci 2008; 28:1865 - 70; 10.1523/JNEUROSCI.5417-07.2008; PMID: 18287503
- Zhang S, Ehlers MD, Bernhardt JP, Su CT, Huganir RL. Calmodulin mediates calcium-dependent inactivation of N-methyl-D-aspartate receptors. Neuron 1998; 21:443 - 53; 10.1016/S0896-6273(00)80553-X; PMID: 9728925
- Ehlers MD, Zhang S, Bernhadt JP, Huganir RL. Inactivation of NMDA receptors by direct interaction of calmodulin with the NR1 subunit. Cell 1996; 84:745 - 55; 10.1016/S0092-8674(00)81052-1; PMID: 8625412
- Mori H, Mishina M. Structure and function of the NMDA receptor channel. Neuropharmacology 1995; 34:1219 - 37; 10.1016/0028-3908(95)00109-J; PMID: 8570021
- Yap KL, Kim J, Truong K, Sherman M, Yuan T, Ikura M. Calmodulin target database. J Struct Funct Genomics 2000; 1:8 - 14; 10.1023/A:1011320027914; PMID: 12836676
- Rhoads AR, Friedberg F. Sequence motifs for calmodulin recognition. FASEB J 1997; 11:331 - 40; PMID: 9141499
- Wang Q, Chen B, Liu P, Zheng M, Wang Y, Cui S, et al. Calmodulin binds to extracellular sites on the plasma membrane of plant cells and elicits a rise in intracellular calcium concentration. J Biol Chem 2009; 284:12000 - 7; 10.1074/jbc.M808028200; PMID: 19254956
- Asano M. Divergent pharmacological effects of three calmodulin antagonists, N-(6-aminohexyl)-5-chloro-1-naphthalenesulfonamide (W-7), chlorpromazine and calmidazolium, on isometric tension development and myosin light chain phosphorylation in intact bovine tracheal smooth muscle. J Pharmacol Exp Ther 1989; 251:764 - 73; PMID: 2810127
- Sun QP, Guo Y, Sun Y, Sun DY, Wang XJ. Influx of extracellular Ca2+ involved in jasmonic-acid-induced elevation of [Ca2+]cyt and JR1 expression in Arabidopsis thaliana.. J Plant Res 2006; 119:343 - 50; 10.1007/s10265-006-0279-x; PMID: 16708291
- Nishikawa M, Hidaka H. Role of calmodulin in platelet aggregation. Structure-activity relationship of calmodulin antagonists. J Clin Invest 1982; 69:1348 - 55; 10.1172/JCI110574; PMID: 7085878
- Sonnhammer EL, von Heijne G, Krogh A. A hidden Markov model for predicting transmembrane helices in protein sequences. Proc Int Conf Intell Syst Mol Biol 1998; 6:175 - 82; PMID: 9783223