Abstract
In a recent study we identified corn cystain9 (CC9) as a novel compatibility factor for the interaction of the biotrophic smut fungus Ustilago maydis with its host plant maize. CC9 is transcriptionally induced during the compatible interaction with U. maydis and localizes in the maize apoplast where it inhibits apoplastic papain-like cysteine proteases. The proteases are activated during incompatible interaction and salicylic acid (SA) treatment and, in turn, are sufficient to induce SA signaling including PR-gene expression. Therefore the inhibition of apoplastic papain-like cysteine proteases by CC9 is essential to suppress host immunity during U. maydis infection. Here were present new experimental data on the cysteine protease-cystatin interaction and provide an in silco analysis of plant cystatins and the identified apoplastic cysteine proteases.
Keywords: :
Within the last years it became evident that cysteine proteases play a pivotal role in programmed cell death and in response to both developmental cues and pathogen attack.Citation1 Plant genomes encode for about 140 cysteine proteases that belong to 15 families.Citation2 Particularly proteases of family C1 Clan CA (papain-like cysteine proteases, PLCPs) are involved in plant-pathogen interactions.Citation1 A prominent example is the infection of tomato by the fungal pathogen Cladosporium fulvum, where the apoplastic PLCP RCR3 is essential for detection of the fungal avirulence protein Avr2 to induce host resistance mediated by the Cf-2 resistance protein.Citation3,Citation4 Silencing of the cysteine protease C14 in tomato results in increased susceptibility to the oomycete Pytophthora infestans.Citation5,Citation6 Beside this, in Nicotiana benthamiana a Cathepsin B participates in the hypersensitive response and resistance to different bacterial pathogens, for example Erwinia amylovora and Pseudomonas syringae.Citation7
Ustilago maydis is a basidiomycetous fungal pathogen that establishes a biotrophic interaction with its host plant maize, leading to the formation of plant tumors in basically all aerial plant organs.Citation8,Citation9 A crucial prerequisite for a successful infection by U. maydis is the suppression of host defense, particularly SA-associated immune responses and cell death. In a previous study we identified the apoplastic corn cystatin9 CC9 being an essential compatibility factor, i.e., transient silencing of cc9 by virus induced gene silencing resulted in host resistance of maize to U. maydis.Citation10 Defense induction in cc9-silenced plants also involves activation of the SA marker genes PR1 and PR5.Citation10 As potential targets for CC9 we identified five apoplastic cysteine proteases of maize, namely CP1A, CP1B, CatB, XCP2 and CP2. Activity of these proteases is strongly activated by SA treatment.Citation10-Citation12 Moreover, infiltration of these proteases was sufficient to induce PR1 expression in naïve plants, indicating a central role of these proteases for the induction of defense responses. In protease activity assays with maize apoplastic fluid, CC9 was able to completely inhibit this SA-induced cysteine protease activity.Citation10 Furthermore, the induction of PR1 was blocked when CC9 was co-infiltrated to the apoplastic cysteine proteases.Citation10 This demonstrates that CC9 is a key compatibility factor during the biotrophic interaction of U. maydis with its host plant maize, since it blocks the activation of SA signaling and further SA dependent defenses responses by inhibition of apoplastic cysteine proteases.Citation10
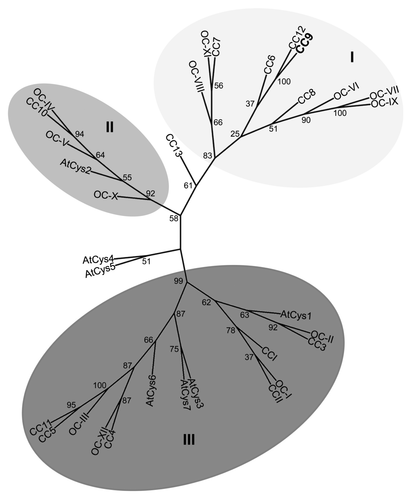
By analyzing the maize genome sequence, 13 different cystatins were identified, which all share the active site QxVxG motif and the phytocystatin consensus sequence ([LVI]-[AGT]-[RKE]-[FY]-[AS]-[VI]-X-[EDQV]-[HYFQ]-N).Citation13-Citation16 In addition to these two common motifs, 11 cystatins harbor an N-terminal signal peptide sequence (in this study we used SignalP 3.0, http://www.cbs.dtu.dk/services/SignalP/). Maize microarray analysis of U. maydis infected seedling leaves revealed that only cc9 is transcriptionally induced during the early biotrophic stage of U. maydis infection.Citation17 A comparison of maize, rice and Arabidopsis cystatins using clustalW2 (http://www.ebi.ac.uk/Tools/msa/clustalw2/) resulted in pairwise scores of amino acid identity between 3% and 89%. The cystatins subdivided into three supported clusters (, phylogenetic analysis was performed based on the clustalW alignments using RAxML BlackBox, http://phylobench.vital-it.ch/raxml-bb/). CC9 belongs to cluster I, which contains only cystatins of the two monocotyledonous plants, while the clusters II and III contain cystatins of all three organisms. The closest homolog of CC9 is CC12, which shows 53% sequence identity but does not contain a predicted secretion signal. By contrast, a phylogenetic analysis did not identify significant homologs of CC9 in A. thaliana or rice, respectively. The Arabidopsis cystatin AtCys1, which was found to be involved in cell death inhibitionCitation18 shares only 16% identity with CC9 and groups into cystatin cluster III ().
Figure 1.Phylogenetic analysis of cystatins from Zea mays, Oryza sativa and Arabidopsis thaliana. Alignment data of full length proteins from Arabidopsis (AtCys), rice (OC) and maize (CC) cystatins were obtained using clustalw2 (http://www.ebi.ac.uk/Tools/msa/clustalw2/). The unrooted tree was calculated using RAxML BlackBox (http://phylobench.vital-it.ch/raxml-bb/) and was visualized by iTOL (http://itol.embl.de/). 100 bootstraps were performed. Bootstrap values are given in the Figure. Bootstrap-supported clusters of related cystatins (cluster I-III) are marked in shades of gray. CC9 is highlighted in bolt letters.
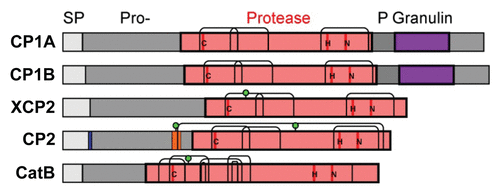
Our previous study showed that CC9 efficiently inhibits the five apoplastic maize PLCPs.Citation10 In silico analysis of these proteins shows that they contain all required features of active proteases: an intact catalytic triad, presence of N-terminal signal peptide indicating secretion, the inhibitory prodomain and a complete protease domain that contains all cysteine pairs that make the conserved disulphide bridges (). The proteases represent four different subfamilies. CP1A and CP1B are very highly similar and differ only in 28 aa. Both belong to subfamily 1 (granulin proteases), which also includes Arabidopsis RD21 and tomato C14.Citation19 Such granulin containing proteases exist in a 40 kDa immature isoform carrying the granulin domain (but lacking the N-terminal prodomain) and a 25 kDa mature isoform.Citation20 XCP2 is a subfamily 3 protease with a putative N-glycosylation site that is unique to this subfamily.Citation19 Proteases of this class, which also contains the well-studied papain, are thought to reside in the xylem. CP2 is a typical aleurain-like protease (subfamily 8) with conserved extra cysteine residues at the end of the prodomain (SRCST) and in the C-terminus (ATCAS).Citation19 An extra cysteine pair makes a disulphide bond that fixes a 10 aa fragment (‘minichain’) of the prodomain in the substrate binding groove. As a result, aleurain proteases have aminopeptidase activity. CP2 also carries aleurain-specific putative N-glycosylation sites in the prodomain and protease domain.Citation19 CP2 also contains the NPIR motif, which is found in over 70% of aleurain-like proteases confers vacuolar targeting.Citation19,Citation21 However, besides CP2 also in tomato, aleurain proteases have been detected in the apoplast.Citation22 The CatB protease belongs to subfamily 9 and carries the three extra cysteine pairs and the conserved putative N-glycosylation site, which is typical for this class of proteases.Citation19 It also contains one additional, unconserved cysteine (, underlined).
Figure 2. Structural features of the apoplastic maize cysteine proteases. All five apoplastic maize PLCPsCitation10 carry a signal peptide (SP) for endomembrane targeting (gray), an autoinhibitory prodomain (Pro-, dark gray), and a protease domain (bright red), which harbours the catalytic triad (C-H-N, red). Two of the proteases (CP1A, CP1B) also carry a C-terminal proline-rich domain (P, gray) and a granulin domain (purple). Putative N-glycosylation sites (green dots) and disulphide bridges (thin bent lines) in the protease domain are indicated. CP2 also carries a vacuolar targeting signal (NPIR, blue) and a minichain (orange) that remains linked to the protease after cleavage of the Pro-domain.
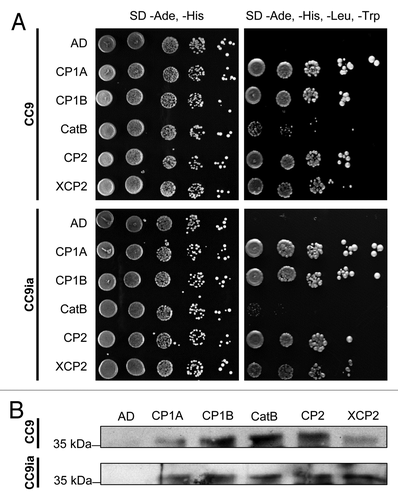
To test for direct interaction of CC9 with the five PLCPs, a Y2H screen was performed (). CC9 or an inactivated form of CC9 (CC9ia)Citation10 were used as bait (binding domain-MYC tag fusion protein). The PLCPs (without secretion signal) served as prey (activation domain-HA tag fusion protein; Matchmaker two-hybride system, Clontech, USA). As a negative control, only the activation domain was co-expressed with CC9 and CC9ia, respectively (). Expression of the individual proteins in yeast was tested by western detection (data not shown). Co-transformation of CC9/CC9ia and all five proteases complemented yeast growth on high stringency plates (SD -Ade,-His,-Leu,-Trp), indicating interaction of CC9 with the proteases (). However, co-expression of CC9 (or CC9ia) with XCP2 and CatB resulted in a reduced growth on high stringency medium when comparing to CP1A, CP1B and CP2. This indicates a weaker interaction between CC9 and these two proteases, particularly in case of CatB.
Figure 3. Interaction of CC9 with the apoplastic maize cysteine proteases by Yeast-2-Hybrid and co-immunoprecipitation. A) Yeast-2-Hybrid interaction assay of CC9 and inactive CC9 (CC9ia) with the five proteases. Yeast drop assays after co- transformation of CC9 (CC9ia) fused to binding domain (BD) with CP1A, CP1B, CatB, CP2 or XCP2 fused to activation domain (AD). Growth on low stringency medium (SD -Ade, -His) selects for expression of AD and BD constructs. Growth of all strains on high stringency medium (SD -Ade, -His, -Leu, -Trp) indicates interaction of the co-expressed proteins. B) Yeast strains shown in (A) were used for co-immunoprecipitation experiments. From each yeast strain, 2 mg of total protein were incubated with 50 µl of anti-HA beads (Roche, Germany) to co-immunoprecipitate the HA-tagged proteases with CC9-MYC. The eluted fractions were analyzed by western blotting using an anti-MYC antibody. CC9-MYC (or CC9ia-MYC) was detected at the expected size (37 kDa), demonstrating protein-protein interaction.
To validate the Y2H data, co-immunoprecipitation assays with total protein extract of the CC9-protease expressing yeast strains were performed (). Using anti-HA matrix, CC9-MYC was co-immunoprecipitated by all tested proteases but not by HA-tagged activation domain, which served as negative control. The same results were obtained when using yeast strains expressing the inactive version of CC9 (CC9ia). These results confirm the observed interaction of CC9 with the five apoplastic cysteine proteases ().
Taken together these data provide further evidence for the interaction of CC9 with the five apoplastic maize PLCPs, although the interaction with XCP2 and, in particular CatB, seems to be less stable when compared with CP1A, CP1B and CP2. This may indicate a minor role of these two proteases during SA dependent defense. In case of XCP2 the interaction with CC9 could also be a byproduct of apoplastic fluid preparation, since this protease is expected to localize mainly in the maize xylem. In addition, the present data suggests that the inactive form of CC9, in which the QxVxG motif is mutated to NxLxA, still interacts with the PLCPs, although CC9ia does not inhibit protease activity.Citation10 This may indicate that further residues outside the QxVxG motif are involved in the cystatin-protease interaction. For the rice cystatin OC-I it was described that a PW-motif in the second hairpin loop also interacts with the active site of the papain,Citation15,Citation23 but this motif is lacking in CC9.
An intriguing question to answer in future approaches is which of the cystatin-protease interactions are actually crucial to modulate plant defense, i.e., to which extent the individual apoplastic PLCP’s contribute to plant immunity.
Sequences
Sequence data of cystatins from rice, A. thaliana and maize CCI-CC10 are published.Citation16,Citation24,Citation25 Sequence data for CC11-CC13 can be found on http://www.maizesequence.org/index.html under the following accession numbers: CC11, GRMZM2G133620; CC12, GRMZM2G312061; CC13, GRMZM2G303361. Sequences of PLCPs are published in.Citation10
| Abbreviations: | ||
| SA | = | salicylic acid |
| JA | = | jasmonic acid |
| CC9 | = | corn cystatin9 |
| CP1A | = | Cysteine protease1-like A |
| CP1B | = | Cysteine protease1-like B |
| CatB | = | CathepsinB3-like |
| XCP-2 | = | Xylem protease2-like |
| CP2 | = | Cysteine protease2-like |
| Y2H | = | yeast two hybrid |
| HA | = | Human influenza hemagglutinin |
| PLCP | = | papain-like cysteine protease |
| PR | = | pathogenesis related |
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
This work was funded by the Max Planck Society and the Deutsche Forschungsgemeinschaft (DFG) via research group FOR 666. For expert help on the phyogenetic analysis of plant cystatins we thank Christine Schauer and Claudia Lüke.
References
- van der Hoorn RAL. Plant proteases: from phenotypes to molecular mechanisms. Annu Rev Plant Biol 2008; 59:191 - 223; 10.1146/annurev.arplant.59.032607.092835; PMID: 18257708
- Rawlings ND, Morton FR, Kok CY, Kong J, Barrett AJ. MEROPS: the peptidase database. Nucleic Acids Res 2008; 36:Database issue D320 - 5; 10.1093/nar/gkm954; PMID: 17991683
- Rooney HC, Van’t Klooster JW, van der Hoorn RAL, Joosten MH, Jones JD, de Wit PJ. Cladosporium Avr2 inhibits tomato Rcr3 protease required for Cf-2-dependent disease resistance. Science 2005; 308:1783 - 6; 10.1126/science.1111404; PMID: 15845874
- van Esse HP, Van’t Klooster JW, Bolton MD, Yadeta KA, van Baarlen P, Boeren S, et al. The Cladosporium fulvum virulence protein Avr2 inhibits host proteases required for basal defense. Plant Cell 2008; 20:1948 - 63; 10.1105/tpc.108.059394; PMID: 18660430
- Tian M, Win J, Song J, van der Hoorn R, van der Knaap E, Kamoun S. A Phytophthora infestans cystatin-like protein targets a novel tomato papain-like apoplastic protease. Plant Physiol 2007; 143:364 - 77; 10.1104/pp.106.090050; PMID: 17085509
- Kaschani F, Shabab M, Bozkurt T, Shindo T, Schornack S, Gu C, et al. An effector-targeted protease contributes to defense against Phytophthora infestans and is under diversifying selection in natural hosts. Plant Physiol 2010; 154:1794 - 804; 10.1104/pp.110.158030; PMID: 20940351
- Gilroy EM, Hein I, van der Hoorn R, Boevink PC, Venter E, McLellan H, et al. Involvement of cathepsin B in the plant disease resistance hypersensitive response. Plant J 2007; 52:1 - 13; 10.1111/j.1365-313X.2007.03226.x; PMID: 17697096
- Skibbe DS, Doehlemann G, Fernandes J, Walbot V. Maize tumors caused by Ustilago maydis require organ-specific genes in host and pathogen. Science 2010; 328:89 - 92; 10.1126/science.1185775; PMID: 20360107
- Schipper K, Doehlemann G. Compatibility in Biotrophic Plant–Fungal Interactions: Ustilago maydis and Friends. In: Perotto S, Baluška F, eds. Signaling and Communication in Plant Symbiosis. Heidelberg, Germany: Springer, 2012:213-38.
- van der Linde K, Hemetsberger C, Kastner C, Kaschani F, van der Hoorn RAL, Kumlehn J, et al. A maize cystatin suppresses host immunity by inhibiting apoplastic cysteine proteases. Plant Cell 2012; 24:1285 - 300; 10.1105/tpc.111.093732; PMID: 22454455
- Doehlemann G, van der Linde K, Assmann D, Schwammbach D, Hof A, Mohanty A, et al. Pep1, a secreted effector protein of Ustilago maydis, is required for successful invasion of plant cells. PLoS Pathog 2009; 5:e1000290; 10.1371/journal.ppat.1000290; PMID: 19197359
- Hemetsberger C, Herrberger C, Zechmann B, Hillmer M, Doehlemann G. The Ustilago maydis effector Pep1 suppresses plant immunity by inhibition of host peroxidase activity. PLoS Pathog 2012; 8:e1002684; 10.1371/journal.ppat.1002684; PMID: 22589719
- Margis R, Reis EM, Villeret V. Structural and phylogenetic relationships among plant and animal cystatins. Arch Biochem Biophys 1998; 359:24 - 30; 10.1006/abbi.1998.0875; PMID: 9799556
- Abraham Z, Martinez M, Carbonero P, Diaz I. Structural and functional diversity within the cystatin gene family of Hordeum vulgare.. J Exp Bot 2006; 57:4245 - 55; 10.1093/jxb/erl200; PMID: 17099080
- Benchabane M, Schlüter U, Vorster J, Goulet MC, Michaud D. Plant cystatins. Biochimie 2010; 92:1657 - 66; 10.1016/j.biochi.2010.06.006; PMID: 20558232
- Massonneau A, Condamine P, Wisniewski JP, Zivy M, Rogowsky PM. Maize cystatins respond to developmental cues, cold stress and drought. Biochim Biophys Acta 2005; 1729:186-99.
- Doehlemann G, Wahl R, Horst RJ, Voll LM, Usadel B, Poree F, et al. Reprogramming a maize plant: transcriptional and metabolic changes induced by the fungal biotroph Ustilago maydis.. Plant J 2008; 56:181 - 95; 10.1111/j.1365-313X.2008.03590.x; PMID: 18564380
- Belenghi B, Acconcia F, Trovato M, Perazzolli M, Bocedi A, Polticelli F, et al. AtCYS1, a cystatin from Arabidopsis thaliana, suppresses hypersensitive cell death. Eur J Biochem 2003; 270:2593 - 604; 10.1046/j.1432-1033.2003.03630.x; PMID: 12787025
- Richau KH, Kaschani F, Verdoes M, Pansuriya TC, Niessen S, Stüber K, et al. Subclassification and biochemical analysis of plant papain-like cysteine proteases displays subfamily-specific characteristics. Plant Physiol 2012; 158:1583 - 99; 10.1104/pp.112.194001; PMID: 22371507
- Yamada K, Matsushima R, Nishimura M, Hara-Nishimura I. A slow maturation of a cysteine protease with a granulin domain in the vacuoles of senescing Arabidopsis leaves. Plant Physiol 2001; 127:1626 - 34; 10.1104/pp.010551; PMID: 11743107
- Ahmed SU, Rojo E, Kovaleva V, Venkataraman S, Dombrowski JE, Matsuoka K, et al. The plant vacuolar sorting receptor AtELP is involved in transport of NH(2)-terminal propeptide-containing vacuolar proteins in Arabidopsis thaliana. J Cell Biol 2000; 149:1335 - 44; 10.1083/jcb.149.7.1335; PMID: 10871276
- Shabab M, Shindo T, Gu C, Kaschani F, Pansuriya T, Chintha R, et al. Fungal effector protein AVR2 targets diversifying defense-related cys proteases of tomato. Plant Cell 2008; 20:1169 - 83; 10.1105/tpc.107.056325; PMID: 18451324
- Nagata K, Kudo N, Abe K, Arai S, Tanokura M. Three-dimensional solution structure of oryzacystatin-I, a cysteine proteinase inhibitor of the rice, Oryza sativa L. japonica. Biochemistry 2000; 39:14753 - 60; 10.1021/bi0006971; PMID: 11101290
- Martinez M, Cambra I, Carrillo L, Diaz-Mendoza M, Diaz I. Characterization of the entire cystatin gene family in barley and their target cathepsin L-like cysteine-proteases, partners in the hordein mobilization during seed germination. Plant Physiol 2009; 151:1531 - 45; 10.1104/pp.109.146019; PMID: 19759340
- Martinez M, Diaz I. The origin and evolution of plant cystatins and their target cysteine proteinases indicate a complex functional relationship. BMC Evol Biol 2008; 8:198; 10.1186/1471-2148-8-198; PMID: 18616807