Abstract
Nicotiana attenuata HSPRO (NaHSPRO) is a negative regulator of seedling growth promoted by the fungus Piriformospora indica. Homologs of NaHSPRO in Arabidopsis thaliana (i.e., AtHSPRO1 and AtHSPRO2) are known to physically interact with the AKINβγ subunit of the SnRK1 complex.Citation2 To investigate whether NaHSPRO is associated with SnRK1 function during the stimulation of seedling growth by P. indica, we studied N. attenuata plants silenced in the expression of NaGAL83 (as-gal83 plants)—a gene that encodes for the regulatory β-subunit of SnRK1—and plants silenced in the expression of both NaHSPRO and NaGAL83 (ir-hspro/as-gal83 plants). The results showed that P. indica differentially stimulated the growth of both as-gal83 and ir-hspro/as-gal83 seedlings compared with control seedlings, with a magnitude similar to that observed in ir-hspro seedlings. Thus, we showed that, similar to NaHSPRO, NaGAL83 is a negative regulator of seedling growth stimulated by P. indica. We propose that the effect of NaHSPRO on seedling growth is associated with SnRK1 signaling.
NaHSPRO from the wild tobacco species Nicotiana attenuata belongs to a group of so-called putative nematode resistance proteins named based on their homology to Hs1pro-1,Citation1 a protein from wild beet (Beta procumbens) that confers resistance to the beet cyst nematode Heterodera schachtii.Citation3 However, the function of Hs1pro-1 homologs (HSPROs) is not only restricted to nematode resistance.Citation3,Citation4 In this regard, HSPROs have also been involved in the regulation of plant defense responses against the bacterial pathogen Pseudomonas syringaeCitation5 and in the regulation of plant mutualistic interactions with the plant growth-promoting fungus Piriformospora indica.Citation1 HSPRO genes have been generically defined as stress responsive genes, and it has been suggested that they play a general role during stress responses.Citation6 However, the molecular function of HSPRO genes remains at present mostly unknown. One important step toward the unraveling of the molecular function of HSPROs has been the discovery that AtHSPRO1 and AtHSPRO2 interact with the AKINβγ regulatory subunit of SnRK1 in Arabidopsis.Citation2 Plant SnRK1s are heterotrimeric protein kinase complexes formed by one catalytic α-subunit (e.g., AKINα10 in Arabidopsis), one regulatory β-subunit (e.g., AKINβ1 in Arabidopsis, NaGAL83 in N. attenuata) and either one regulatory γ- or one chimeric βγ-subunit (AKINγ1 and AKINβγ in Arabidopsis, respectively).Citation2,Citation7 It has been reported that silencing either the expression of the catalytic α-subunit or the regulatory β-subunit of plant SnRK1s leads to an inactive complex.Citation8 In all organisms studied thus far, SnRK1s sense the energy status of the cells, and in plants, SnRK1 represses the expression of genes involved in energy-consuming biosynthetic processes and activates the expression of catabolic genes responsible for increasing energy and nutrient availability under conditions in which those are scarce.Citation9 In addition to the regulation of gene expression, SnRK1 functions as a regulator of the activity of key nitrogen and carbon metabolic enzymesCitation6,Citation10 and the transport of photo-assimilates from shoots to roots during tolerance responses to insect herbivory.Citation3 Recently, it has been shown that Arabidopsis and rice (Oriza sativa) SnRK1s participate in stress-responsive gene regulation and in plant growth and development.Citation11
Considering the previously published study on the interaction between AKINβγ and AtHSPRO1 and AtHSPRO2 in Arabidopsis,Citation2 and the fact that this interaction occurs in the cytosol (coincident with the cellular localization of NaHSPRO in N. attenuata),Citation1 we hypothesized that changes in metabolism brought about by SnRK1-mediated mechanisms underlie the differential growth promotion of ir-hspro seedlings during the interaction with P. indica.Citation1 To test this hypothesis, we followed a genetic approach in which we assessed seedling growth in the progeny derived from the following crosses (performed reciprocally for each genotype): WT x ir-hspro (two independent transgenic lines were used: ir-hspro 1 and ir-hspro 2),Citation1 WT x as-gal83 (a transgenic N. attenuata line stably silenced in the expression of NaGAL83 by antisense technique)Citation7 and ir-hspro x as-gal83. The crosses between singly silenced lines and WT plants were performed as a control for the cross between ir-hspro and as-gal83 plants and therefore for hemizygosity of the transgenes. An additional cross between WT and a transgenic N. attenuata line stably silenced in the expression of the transcription factor HD20 (ir-hd20)Citation3 was used as a transgenic-plant control. WT plants were also manually self-crossed as a control (WT x WT). Seedling fresh weight of all genotypes used was quantified after 14 d of growth in both the presence and absence of P. indica and the assay was conducted in a plate system as previously described.Citation1 As expected, increased growth promotion (45% gain in fresh biomass) was observed in WT seedlings grown in the presence of P. indica as compared with WT seedlings grown in the absence of the fungus ( and ) and a stronger growth promotion (ca. 250%) was observed for WT x ir-hspro seedlings grown under the same conditions ( and ). WT x as-gal83 and ir-hspro/as-gal83 seedlings showed a growth promotion effect of the same magnitude as that quantified for WT x ir-hspro seedlings in the presence of P. indica ( and ). In contrast, the growth of WT x ir-hd20 seedlings was not differentially stimulated by the fungus compared with WT seedlings ( and ). The results showed that, during interaction with P. indica, the silencing of NaGAL83 expression (either alone or in combination with NaHSPRO) produces the same effect on seedling growth as the silencing of NaHSPRO alone. From a genetic perspective, these results strongly suggested that these two genes act in the same pathway and therefore that HSPRO negatively regulates WT N.attenuata seedling growth during interaction with P. indica by affecting SnRK1-mediated signaling.
Figure 1. Differential growth promotion of Nicotiana attenuata seedlings mediated by Piriformospora indica. After seven days of germination, N. attenuata seedlings were transferred onto PNM (plant nutrient medium)Citation1 and inoculated with P. indica culture or a medium control. After 14 d of incubation in these conditions, the fresh biomass of seedlings was determined with a microbalance (n = 8–17; bars denote standard error of the mean). Statistical analysis was conducted using one way-ANOVA with Tuckey post-hoc test. The letters on top of the bars denote significant differences between genotypes and treatments and the P-values associated with the analysis are summarized in .
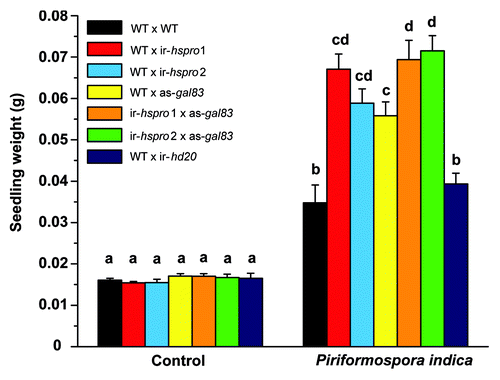
Table 1. One way-ANOVA and Tuckey post-hoc test results from the comparison of Piriformospora indica-colonized seedlings presented in
SnRK1 is activated under stress conditions such as nutrient and energy deprivation, playing a central role in metabolic and transcriptional reprogramming of processes necessary for the switch in resource utilization and allocation and affecting thereby growth.Citation6 In cases where SnRK1 activity is inhibited (e.g., exogenous supply of sugars), plant growth is promoted. As an example, silencing the expression of AKINα10 leads to enhanced Arabidopsis seedling growth via a more efficient use of exogenously supplied sucrose and glucose.Citation9 In N. attenuata plants, repression of NaGAL83 expression (and thereby inhibition of SnRK1 activity) during insect herbivory induces an increased transport of photo-assimilates from shoots to roots (C “bunkering”), thus delaying senescence and prolonging flowering.Citation3
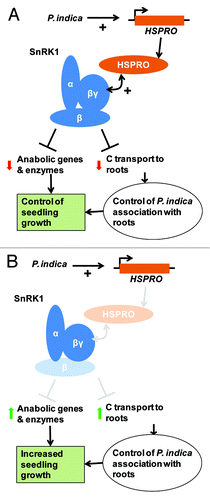
Taking all results together, we propose two possible scenarios (non-mutually exclusive) to describe the role of HSPRO and SnRK1 in controlling WT N. attenuata seedling growth during P. indica interaction. In one scenario, activation of HSPRO expression by the interaction of roots with P. indica subsequently activates SnRK1 (by still unknown mechanisms but based on the interaction of HSPRO with the regulatory βγ-subunit of SnRK1). SnRK1 activation inhibits increased photo-assimilate transport from the shoot to the root, controlling root sugar content and the beneficial interaction with P. indica (i.e., stimulation of seedling growth; ). In the cases where the activity of SnRK1 is reduced (e.g., as-gal83 plants) or the expression of HSPRO is impaired (e.g., ir-hspro plants), changes in sugar content in roots affect the beneficial interaction of P. indica with this tissue in a manner that induces differential growth promotion of seedlings (). It is important to note that colonization rates of P. indica are similar between ir-hspro plants and WT,Citation1 and therefore the hypothetical changes in beneficial interaction are independent of higher colonization rates by the fungus. This scenario assumes that wild-type rates of C allocation in roots are an important determinant of the interaction of this tissue with P. indica. This assumption is consistent with previous studies showing that, in P. indica colonized barley roots, fungal proteins involved in carbon and nitrogen uptake and metabolism are important for the switch from a biotrophic to a necrotrophic lifestyle by the fungus.Citation13 Another study performed with Arabidopsis and tobacco (N. tabacum) plants showed that the expression of plant enzymes involved in starch degradation and nitrate assimilation is induced in P. indica-colonized roots.Citation14 In a second scenario, activation of HSPRO expression by the interaction of roots with P. indica activates SnRK1, and this activation leads to a more direct negative feedback control on seedling growth, for example, by repression of biosynthesis genes or deactivation of biosynthesis enzymes (). When either HSPRO or SnRK1 activities or expression are lower (e.g., in ir-hspro or as-gal83 plants) the negative feedback on seedling growth is less restrained and seedlings increase their growth rates (). It is noteworthy that in tomato (Solanum lycopersicum), the β-subunit of SnRK1 (the ortholog of NaGAL83) is phosphorylated by the protein kinase Adi3 (AvrPto-dependent Pto-interacting protein3).Citation15 Thus, by associating with the βγ-subunit of SnRK1, HSPRO could interfere with the post-translation modification of, for example, NaGAL83 in N. attenuata.
Figure 2. Hypothetical model for the role of HSPRO and SnRK1 in regulation of seedling growth during interaction of N. attenuata plants with the growth promoting fungus P. indica.(A) The interaction of P. indica with roots of N. attenuata seedlings induces the expression of HSPRO via transcriptional activation.Citation1 HSPRO interacts with the βγ-subunit of SnRK1Citation2 and this interaction activates SnRK1 by still unknown mechanisms. Activation of SnRK1 controls the allocation of carbon (C) from photo-assimilates to the rootsCitation3 and negatively regulates the expression of anabolic genes and enzymes (references in text). These mechanisms either directly or indirectly affect seedling growth during interaction with P. indica. (B) In plants with reduced levels of HSPRO expression (e.g., triggered by gene silencing) or SnRK1 activity (e.g., triggered by silencing of the β-subunit), the negative regulation of C transport to roots, anabolic gene expression and metabolic enzyme activities by SnRK1 is lessened and seedling growth is enhanced via differential growth promotion by P. indica.
In conclusion, the experiments presented in this study provided strong evidence for the participation of HSPRO in the regulation of SnRK1-mediated responses controlling N. attenuata seedling growth during interaction with P. indica. Interestingly, the effect of differential growth promotion of ir-hspro and as-gal83 seedlings only occurs during interaction with P. indica, indicating that activation of HSPRO/SnRK1 by this fungus is required. New exciting hypothesis have been derived from these experiments and future studies will be focused on the understanding of the molecular connection between HSPRO and SnRK1 and on how this connection regulates metabolism to accelerate seedling growth during interaction with P. indica and other growth promoting microorganisms.
| Abbreviations: | ||
| Adi3 | = | AvrPto-dependent Pto-interacting protein 3 (Solanum lycopersicum) |
| AKINα10, SNF1 KINASE HOMOLOG 10, synonym | = | AKINα2 (Arabidopsis thaliana) |
| AKINβγ, HOMOLOG OF YEAST SUCROSE NONFERMENTING 4, synonym | = | SNF1-related protein kinase regulatory subunit βγ (Arabidopsis thaliana) |
| AKINγ1, SNF1-RELATED PROTEIN KINASE REGULATORY SUBUNIT γ 1, synonym | = | SNF1-related protein kinase regulatory subunit gamma-1 (Arabidopsis thaliana) |
| as | = | antisense |
| GAL83 | = | GALactose metabolism mutant 83 (Saccharomyces cerevisiae) |
| HD20 | = | homeodomain 20 transcription factor (Nicotiana attenuata) |
| HSPRO | = | ortholog of nematode resistance protein Hs1pro-1 from Beta procumbens |
| ir | = | inverted-repeat |
| SNF1 | = | sucrose non-fermenting-1 (Saccharomyces cerevisiae) |
| SnRK1 | = | SNF1-related protein kinase 1 |
Acknowledgments
This study was financially supported by the Deutsche Forschungsgesellschaft (DFG; BO3260/3-1 and 3-2) and the Max Planck Society.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- Schuck S, Camehl I, Gilardoni PA, Oelmueller R, Baldwin IT, Bonaventure G. HSPRO controls early Nicotiana attenuata seedling growth during interaction with the Fungus Piriformospora indica.. Plant Physiol 2012; 160:929 - 43; http://dx.doi.org/10.1104/pp.112.203976; PMID: 22892352
- Gissot L, Polge C, Jossier M, Girin T, Bouly JP, Kreis M, et al. AKINbetagamma contributes to SnRK1 heterotrimeric complexes and interacts with two proteins implicated in plant pathogen resistance through its KIS/GBD sequence. Plant Physiol 2006; 142:931 - 44; http://dx.doi.org/10.1104/pp.106.087718; PMID: 17028154
- Cai D, Kleine M, Kifle S, Harloff HJ, Sandal NN, Marcker KA, et al. Positional cloning of a gene for nematode resistance in sugar beet. Science 1997; 275:832 - 4; http://dx.doi.org/10.1126/science.275.5301.832; PMID: 9012350
- McLean MD, Hoover GJ, Bancroft B, Makhmoudova A, Clark SM, Welacky T, et al. Identification of the full-length Hs1pro-1 coding sequence and preliminary evaluation of soybean cyst nematode resistance in soybean transformed with Hs1pro-1 cDNA. Canadian Journal of Botany-Revue Canadienne De Botanique 2007; 85:437 - 41; http://dx.doi.org/10.1139/B07-038
- Murray SL, Ingle RA, Petersen LN, Denby KJ. Basal resistance against Pseudomonas syringae in Arabidopsis involves WRKY53 and a protein with homology to a nematode resistance protein. Mol Plant Microbe Interact 2007; 20:1431 - 8; http://dx.doi.org/10.1094/MPMI-20-11-1431; PMID: 17977154
- Baena-González E, Sheen J. Convergent energy and stress signaling. Trends Plant Sci 2008; 13:474 - 82; http://dx.doi.org/10.1016/j.tplants.2008.06.006; PMID: 18701338
- Schwachtje J, Minchin PEH, Jahnke S, van Dongen JT, Schittko U, Baldwin IT. SNF1-related kinases allow plants to tolerate herbivory by allocating carbon to roots. Proc Natl Acad Sci USA 2006; 103:12935 - 40; http://dx.doi.org/10.1073/pnas.0602316103; PMID: 16912118
- Halford NG, Hey S, Jhurreea D, Laurie S, McKibbin RS, Paul M, et al. Metabolic signalling and carbon partitioning: role of Snf1-related (SnRK1) protein kinase. J Exp Bot 2003; 54:467 - 75; http://dx.doi.org/10.1093/jxb/erg038; PMID: 12508057
- Baena-González E, Rolland F, Thevelein JM, Sheen J. A central integrator of transcription networks in plant stress and energy signalling. Nature 2007; 448:938 - 42; http://dx.doi.org/10.1038/nature06069; PMID: 17671505
- Sugden C, Donaghy PG, Halford NG, Hardie DG. Two SNF1-related protein kinases from spinach leaf phosphorylate and inactivate 3-hydroxy-3-methylglutaryl-coenzyme A reductase, nitrate reductase, and sucrose phosphate synthase in vitro. Plant Physiol 1999; 120:257 - 74; http://dx.doi.org/10.1104/pp.120.1.257; PMID: 10318703
- Cho YH, Hong JW, Kim EC, Yoo SD. Regulatory functions of SnRK1 in stress-responsive gene expression and in plant growth and development. Plant Physiol 2012; 158:1955 - 64; http://dx.doi.org/10.1104/pp.111.189829; PMID: 22232383
- Ré DA, Raud B, Chan RL, Baldwin IT, Bonaventure G. RNAi-mediated silencing of the HD-Zip gene HD20 in Nicotiana attenuata affects benzyl acetone emission from corollas via ABA levels and the expression of metabolic genes. BMC Plant Biol 2012; 12:60; http://dx.doi.org/10.1186/1471-2229-12-60; PMID: 22548747
- Zuccaro A, Lahrmann U, Güldener U, Langen G, Pfiffi S, Biedenkopf D, et al. Endophytic life strategies decoded by genome and transcriptome analyses of the mutualistic root symbiont Piriformospora indica.. PLoS Pathog 2011; 7:e1002290; http://dx.doi.org/10.1371/journal.ppat.1002290; PMID: 22022265
- Sherameti I, Shahollari B, Venus Y, Altschmied L, Varma A, Oelmüller R. The endophytic fungus Piriformospora indica stimulates the expression of nitrate reductase and the starch-degrading enzyme glucan-water dikinase in tobacco and Arabidopsis roots through a homeodomain transcription factor that binds to a conserved motif in their promoters. J Biol Chem 2005; 280:26241 - 7; http://dx.doi.org/10.1074/jbc.M500447200; PMID: 15710607
- Avila J, Gregory OG, Su DY, Deeter TA, Chen SX, Silva-Sanchez C, et al. The β-subunit of the SnRK1 complex is phosphorylated by the plant cell death suppressor Adi3. Plant Physiol 2012; 159:1277 - 90; http://dx.doi.org/10.1104/pp.112.198432; PMID: 22573803