Abstract
Xyloglucan endotransglucosylase, catalyzed by XTH subfamily members, is thought to play crucial roles in plant cell wall physiology. Recent discovery of endotransglycosylases active on other hemicelluloses extend our understanding of the physiological scope of endotransglycosylation in general. Discovery in Poaceaen XTHs of endotransglycosylases which act on Poaceaen-prevalent hemicelluloses, such as MLG, could reconcile the apparent incongruence between the large size of Poaceaen putative XTH families and the low xyloglucan content of their cell walls. Here, I speculate on hypothetical MLG-active endotransglycosylases and highlight potential hindrances to their discovery. It is suggested that because the location of β-(1→3) bonds within MLG oligosaccharides (MLGOs) could define their ability to act as endotranglycosylase acceptor substrates: a) thorough probing of substrate specificities necessitates the use of MLGOs created using different endo-glycanases; and b) endogenous plant exo-glycosidases, which can hinder endotranglycosylase assays by degrading acceptor substrates, might prove particularly troublesome where MLGOs are concerned.
The plant cell wall is a hydrated extraprotoplasmic complex which plays crucial roles in plant life.Citation1 It is composed largely of polysaccharides: cellulose, hemicellulose and pectin. The presence and fine structure of hemicelluloses vary in a phylogenetically-dependent manner.Citation2-Citation5 For example, within land plants, xyloglucan appears ubiquitousCitation4,Citation5 while mixed-linkage (1→3),(1→4)-β-d-glucan (MLG) is known only in the PoalesCitation5,Citation6 and Equisetum.Citation7,Citation8 Xyloglucan is a β-(1→4)-d-glucan backbone with additional glycosyl substitutions.Citation3,Citation9 MLG is an unbranched d-glucosyl homopolymer in which straight β-(1→4)-linked regions, typically 3–4 residues, are connected by single β-(1→3) bonds which cause kinks in the structureCitation10 ().
Figure 1. Endotransglycosylase reactions and the use of MLGOs as acceptor substrates. (A) MLG polymer structure showing kinks caused by β-(1→3) bonds as well as distinct sites of enzymic attack: lichenase cleaves β-(1→4) bonds after β-(1→3) bondsCitation29 (gray downward triangles), cellulase cleaves β-(1→4) bonds before β-(1→3) bondsCitation30 (black upward triangles); (B) General endotransglycosylase reaction. During assays the acceptor substrate is typically a labeled (e.g., radioactive) oligosaccharide. Labeled polymeric material is detected after the reaction; Major products of lichenase (C) and cellulase (D) digestion of MLG as each might fit into XTH positive (acceptor substrate-binding) subsites; (E) 5 successive β-D-glucosidase digestion reactions (each releasing Glc from the non-reducing terminus) on a MLG-hexasaccharide (produce by incomplete lichenase digestion) demonstrating the alternation of position of the β-(1→3) kink relative to XTH positive subsites. Reducing termini on right; blue squares, D-Glc residues; β3, β-(1→3)-linkage (highlighted in orange); β4, β-(1→4)-linkage; cello-like β-(1→4)-linked regions underlined.
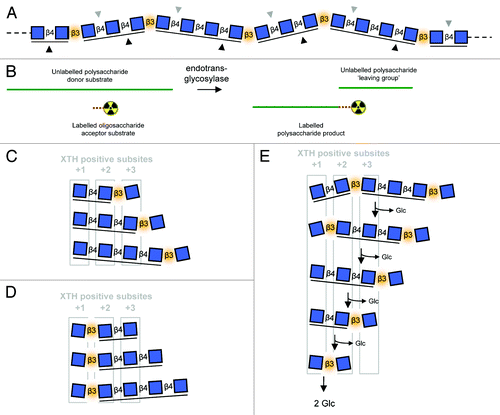
Endotransglycosylases catalyze the endo-cleavage of a glycan (the donor substrate) and the creation of a glycosidic link between the newly formed potentially reducing terminus and the non-reducing terminus of another glycan (the acceptor substrate; ). Xyloglucan endotransglucosylase (XET) activityCitation11,Citation12—which uses xyloglucan both as donor and acceptor—is catalyzed by xyloglucan endotransglucosylase/hydrolase (XTH) subfamily members, appears ubiquitous in the land plants and is believed to play crucial roles in cell wall physiology.Citation13 XTH active sites contain seven subsites (−4, −3, −2, −1, +1, +2, +3), each binding a xyloglucan backbone residue and/or its substitutions; donor cleavage occurs between subsites −1 and +1, before the acceptor substrate occupies the positive subsites.Citation14
The relative recentness of the discovery of other endotransglycosylase activitiesCitation15-Citation18 is perhaps surprising given the crucial role thought to be played by XET and the wide diversity of hemicelluloses within the plant kingdom. The apparent incongruity between the low xyloglucan content of Poaceaen cell wallsCitation3,Citation19 and the large number of putative XTH genes they maintainCitation20 (e.g., 30 and 32 in rice and maize, respectively)Citation14 could be reconciled if some were shown to catalyze significant levels of non-XET endotransglycosylase reaction/s.Citation21 As MLG is one of the most prevalent hemicelluloses, as well as a distinctive feature, of the “Type II” cell walls possessed by the Poaceae,Citation2,Citation3,Citation5,Citation19 an MLG-active endotransglycosylase—such as mixed-linkage (1→3),(1→4)-β-d-glucan: xyloglucan endotransglucosylase (MXE), which uses MLG as a donor and xyloglucan as an acceptorCitation16—would be a reasonable focus of attention. However, the only reports of MXE and MXE-like activities outside Equisetum were “side reactions” catalyzed by predominantly XET-active enzymes.Citation22,Citation23 Recently a novel method to assay endotransglycosylase “action” in vivo (compare “activity” in vitro) was devised,Citation24 in which [3H]xyloglucan oligosaccharitols ([3H]XGO-ols) permeated the cell walls of intact living plant tissues, allowing them to be acted on in situ by endogenous enzymes with endogenous donor substrates. Subsequent enzymic digestion of any resulting [3H]polymers identified the donor substrate (e.g., digestion with lichenase—“MLG-endoglucanase”—indicated MXE). This method showed speculation that Poacean cell walls might contain inextractable MXECitation16,Citation22 to be untrue.
Despite these evidences against MXE in the Poaceae, there still remains huge scope for other potential MLG-active endotransglycosylases, which could theoretically use any donor: acceptor combination conceivable. The novel in vivo method described above can stand alongside previously devised high-throughput screens,Citation25,Citation26 all of which can facilitate the testing of this plethora of combinations. However, digestion of candidate acceptor substrates by endogenous plant exo-glycosidases is a constant threat to such assays, all of which use un-purified enzymes. [3H]XGO-ol resistance to β-D-glucosidase makes them particularly amenable to these methods, but candidate acceptors with free non-reducing terminal β-d-glucoses, such as MLG oligosaccharides (MLGOs), are highly prone to it,Citation17 potentially rendering them too short to participate in endotransglycosylation. Accommodation or tackling of this might prove vital for identification of predominantly MLG-active endotransglycosylases: because all sequenced endotransglycosylases to dateCitation18,Citation27,Citation28 are homo-endotransglycosylases (use same glycan type as donor and acceptor), a hypothetical endotransglycosylase using MLG both as donor and acceptor is perhaps a more likely candidate for an MLG-active endotransglycosylase than MXE. Some other endotransglycosylase activities using MLGOs as acceptors have been observed, though for now only as side-reactions of predominantly XET-active enzymesCitation23 or minor activities in crude preparations.Citation26
A vital consideration for the design of endotransglycosylase assays using MLGOs as acceptors is that, just as MLG-hydrolysing enzymes typically cleave at specific linkages relative to β-(1→3) bonds (, lichenase,Citation29 cellulaseCitation30), it is highly likely that an endotransglycosylase capable of utilizing MLGOs as acceptors will exhibit a similar preference for the position of the β-(1→3) bond relative to their non-reducing termini (and thus the enzyme’s catalytic residues). Given that the identity of such a preference is currently unknown, use of MLGOs created by digestion with different enzymes () would be crucial to enable the full probing of potential MLGO-utilizing endotransglycosylases. If the active site of a hypothetical MLGO-utilizing XTH-homolog is shaped to suit a β-(1→3) kink between subsites +1 and +2, it will favor cellulase-produced MLGOs. Probing substrate specificities with lichenase-produced MLGOs aloneCitation23,Citation26 might overlook some MLG-active endotransglycosylases. Furthermore, because of this, endogenous β-D-glucosidases would confound the use of MLGOs 2-fold: as well as shortening them, the resultant constant interchange of β-(1→3) bond position relative to the non-reducing terminus means that, even when still of sufficient length, MLGOs might rarely be suitable substrates (). The scope for future work is both encouraging and perhaps daunting. But the consideration of these issues will be necessary before the presence and prevalence of speculated MLG-active endotransglycosylases can be fully assessed.
| Abbreviations: | ||
| MLG | = | mixed-linkage (1→3),(1→4)-β-d-glucan |
| XET | = | xyloglucan endotransglucosylase |
| XTH | = | xyloglucan endotransglucosylase/hydrolase |
| MXE, mixed-linkage (1→3),(1→4)-β-d-glucan | = | xyloglucan endotransglucosylase |
| XGO-ol | = | xyloglucan oligosaccharitol |
Acknowledgments
I thank the BBSRC for a PhD studentship.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- Albersheim P, Darvill A, Roberts K, Sederoff R, Staehelin A. Plant cell walls. New York, NY: Garland Science, 2011.
- Popper ZA. Evolution and diversity of green plant cell walls. Curr Opin Plant Biol 2008; 11:286 - 92; http://dx.doi.org/10.1016/j.pbi.2008.02.012; PMID: 18406657
- Scheller HV, Ulvskov P. Hemicelluloses. Annu Rev Plant Biol 2010; 61:263 - 89; http://dx.doi.org/10.1146/annurev-arplant-042809-112315; PMID: 20192742
- Popper ZA, Fry SC. Primary cell wall composition of bryophytes and charophytes. Ann Bot 2003; 91:1 - 12; http://dx.doi.org/10.1093/aob/mcg013; PMID: 12495914
- Popper ZA, Fry SC. Primary cell wall composition of pteridophytes and spermatophytes. . New Phytol 2004; 164:165 - 74; http://dx.doi.org/10.1111/j.1469-8137.2004.01146.x
- Smith BG, Harris PJ. The polysaccharide composition of Poales cell walls: Poaceae cell walls are not unique. Biochem Syst Ecol 1999; 27:33 - 53; http://dx.doi.org/10.1016/S0305-1978(98)00068-4
- Fry SC, Nesselrode BHWA, Miller JG, Mewburn BR. Mixed-linkage (1-->3,1-->4)-β-D-glucan is a major hemicellulose of Equisetum (horsetail) cell walls. New Phytol 2008; 179:104 - 15; http://dx.doi.org/10.1111/j.1469-8137.2008.02435.x; PMID: 18393951
- Sørensen I, Pettolino FA, Wilson SM, Doblin MS, Johansen B, Bacic A, et al. Mixed-linkage (1→3,1→4)-β-D-glucan is not unique to the Poales and is an abundant component of Equisetum arvense cell walls. Plant J 2008; 54:510 - 21; http://dx.doi.org/10.1111/j.1365-313X.2008.03453.x; PMID: 18284587
- Fry SC. The Structure and Functions of Xyloglucan. J Exp Bot 1989; 40:1 - 11; http://dx.doi.org/10.1093/jxb/40.1.1
- Burton RA, Fincher GB. (1,3;1,4)-β-D-glucans in cell walls of the poaceae, lower plants, and fungi: a tale of two linkages. Mol Plant 2009; 2:873 - 82; http://dx.doi.org/10.1093/mp/ssp063; PMID: 19825664
- Fry SC, Smith RC, Renwick KF, Martin DJ, Hodge SK, Matthews KJ. Xyloglucan endotransglycosylase, a new wall-loosening enzyme activity from plants. Biochem J 1992; 282:821 - 8; PMID: 1554366
- Nishitani K, Tominaga R. Endo-xyloglucan transferase, a novel class of glycosyltransferase that catalyzes transfer of a segment of xyloglucan molecule to another xyloglucan molecule. J Biol Chem 1992; 267:21058 - 64; PMID: 1400418
- Rose JK, Braam J, Fry SC, Nishitani K. The XTH family of enzymes involved in xyloglucan endotransglucosylation and endohydrolysis: current perspectives and a new unifying nomenclature. Plant Cell Physiol 2002; 43:1421 - 35; http://dx.doi.org/10.1093/pcp/pcf171; PMID: 12514239
- Eklöf JM, Brumer H. The XTH gene family: an update on enzyme structure, function, and phylogeny in xyloglucan remodeling. Plant Physiol 2010; 153:456 - 66; http://dx.doi.org/10.1104/pp.110.156844; PMID: 20421457
- Schröder R, Wegrzyn TF, Bolitho KM, Redgwell RJ. Mannan transglycosylase: a novel enzyme activity in cell walls of higher plants. Planta 2004; 219:590 - 600; http://dx.doi.org/10.1007/s00425-004-1274-x; PMID: 15118857
- Fry SC, Mohler KE, Nesselrode BHWA, Franková L. Mixed-linkage beta-glucan : xyloglucan endotransglucosylase, a novel wall-remodelling enzyme from Equisetum (horsetails) and charophytic algae. Plant J 2008; 55:240 - 52; http://dx.doi.org/10.1111/j.1365-313X.2008.03504.x; PMID: 18397375
- Franková L, Fry SC. Phylogenetic variation in glycosidases and glycanases acting on plant cell wall polysaccharides, and the detection of transglycosidase and trans-β-xylanase activities. Plant J 2011; 67:662 - 81; http://dx.doi.org/10.1111/j.1365-313X.2011.04625.x; PMID: 21554451
- Johnston SL, Prakash R, Chen NJ, Kumagai MH, Turano HM, Cooney JM, et al. An enzyme activity capable of endotransglycosylation of heteroxylan polysaccharides is present in plant primary cell walls. Planta 2013; 237:173 - 87; http://dx.doi.org/10.1007/s00425-012-1766-z; PMID: 23001197
- Carpita NC, Gibeaut DM. Structural models of primary cell walls in flowering plants: consistency of molecular structure with the physical properties of the walls during growth. Plant J 1993; 3:1 - 30; http://dx.doi.org/10.1111/j.1365-313X.1993.tb00007.x; PMID: 8401598
- Yokoyama R, Rose JKC, Nishitani K. A surprising diversity and abundance of xyloglucan endotransglucosylase/hydrolases in rice. Classification and expression analysis. Plant Physiol 2004; 134:1088 - 99; http://dx.doi.org/10.1104/pp.103.035261; PMID: 14988479
- Strohmeier M, Hrmova M, Fischer M, Harvey AJ, Fincher GB, Pleiss J. Molecular modeling of family GH16 glycoside hydrolases: potential roles for xyloglucan transglucosylases/hydrolases in cell wall modification in the poaceae. Protein Sci 2004; 13:3200 - 13; http://dx.doi.org/10.1110/ps.04828404; PMID: 15557263
- Hrmova M, Farkaš V, Lahnstein J, Fincher GB. A Barley xyloglucan xyloglucosyl transferase covalently links xyloglucan, cellulosic substrates, and (1,3;1,4)-β-D-glucans. J Biol Chem 2007; 282:12951 - 62; http://dx.doi.org/10.1074/jbc.M611487200; PMID: 17329246
- Stratilová E, Ait-Mohand F, Rehulka P, Garajová S, Flodrová D, Rehulková H, et al. Xyloglucan endotransglycosylases (XETs) from germinating nasturtium (Tropaeolum majus) seeds: isolation and characterization of the major form. Plant Physiol Biochem 2010; 48:207 - 15; http://dx.doi.org/10.1016/j.plaphy.2010.01.016; PMID: 20153658
- Mohler KE, Simmons TJ, Fry SC. Mixed-linkage glucan:xyloglucan endotransglucosylase (MXE) re-models hemicelluloses in Equisetum shoots but not in barley shoots or Equisetum callus. New Phytol 2013; 197:111 - 22; http://dx.doi.org/10.1111/j.1469-8137.2012.04371.x; PMID: 23078260
- Fry SC. Novel ‘dot-blot’ assays for glycosyltransferases and glycosylhydrolases: optimization for xyloglucan endotransglycosylase (XET) activity. Plant J 1997; 11:1141 - 50; http://dx.doi.org/10.1046/j.1365-313X.1997.11051141.x
- Kosík O, Auburn RP, Russell S, Stratilová E, Garajová S, Hrmova M, et al. Polysaccharide microarrays for high-throughput screening of transglycosylase activities in plant extracts. Glycoconj J 2010; 27:79 - 87; http://dx.doi.org/10.1007/s10719-009-9271-8; PMID: 19953317
- Okazawa K, Sato Y, Nakagawa T, Asada K, Kato I, Tomita E, et al. Molecular cloning and cDNA sequencing of endoxyloglucan transferase, a novel class of glycosyltransferase that mediates molecular grafting between matrix polysaccharides in plant cell walls. J Biol Chem 1993; 268:25364 - 8; PMID: 8244968
- Schröder R, Wegrzyn TF, Sharma NN, Atkinson RG. LeMAN4 endo-β-mannanase from ripe tomato fruit has dual enzyme activity and can act as a mannan transglycosylase and hydrolase. Planta 2006; 224:1091 - 102; http://dx.doi.org/10.1007/s00425-006-0286-0; PMID: 16649044
- Planas A. Bacterial 1,3-1,4-β-glucanases: structure, function and protein engineering. Biochim Biophys Acta 2000; 1543:361 - 82; http://dx.doi.org/10.1016/S0167-4838(00)00231-4; PMID: 11150614
- Grishutin SG, Gusakov AV, Dzedzyulya EI, Sinitsyn AP. A lichenase-like family 12 endo-(1-->4)-β-glucanase from Aspergillus japonicus: study of the substrate specificity and mode of action on β-glucans in comparison with other glycoside hydrolases. Carbohydr Res 2006; 341:218 - 29; http://dx.doi.org/10.1016/j.carres.2005.11.011; PMID: 16343463