Abstract
The vesicle inducing protein in plastids (VIPP1) is an essential protein for the biogenesis of thylakoids in modern cyanobacteria, algae, and plants. Although its exact function is still not clear, recent work has provided important hints to its mode of action. We believe that these data are consistent with a structural role of VIPP1 within thylakoid centers, which are considered as sites from which thylakoid membranes emerge and at which the biogenesis at least of photosystem II is thought to occur. Here we present a model that may serve as starting point for future research.
The structure and function of the major photosynthetic complexes in thylakoid membranes are well characterized.Citation1 However, the biogenesis of thylakoid membranes and of most of the major complexes it harbors is barely understood.Citation2 This is surprising, as the thylakoid membranes are the basis for almost all life on earth. An important role in the biogenesis of thylakoid membranes, but not of the protein complexes it harbors, was suggested for the vesicle inducing proteins in plastids (VIPP1).Citation3-Citation5 However, this assignment was recently challenged by several reports.Citation6-Citation10 Based on current knowledge, we will in the following construct a model for a role of VIPP1 as a dynamic, structural component of thylakoid centers.
In cyanobacteria, the biogenesis at least of photosystem (PS) II appears to take place at distinct sites referred to as thylakoid centers.Citation11,Citation12 Thylakoid centers are located close to the cytoplasma membrane and are situated at the origin of multiple thylakoid membrane layersCitation13,Citation14 (). They contain rod-shaped structures of 50- to 1000-nm length and 30- to 50-nm width that consist of ring-like structures with ~14-fold symmetry ( and ). Similar structures, referred to as microtubule-like structures (MTLs), have been observed in early electron microscopy studies in plastids of diverse algal and plant speciesCitation15-Citation18 (). MTLs have diameters of 15 to 40 nm, reach lengths of up to 5 µm, and appear to bridge inner envelope and thylakoid membranes.
Figure 1. Features of cyanobacterial thylakoid centers. (A) Thylakoids radiating from 2 thylakoid centers (arrows) forming an interconnection between 2 centers in Pleurocapsa minor (bar = 50 nm). (B) Longitudinal section of a thylakoid center (arrow) in Anabaena cylindrica (bar = 50 nm). (C) Markham rotation enhancement of a thylakoid center from Dermocarpa violaceae. The distinct 14-subunit pattern is marked with arrows. Adopted from Kunkel (1982).Citation13
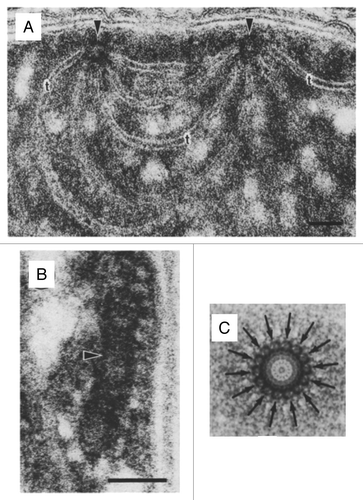
Figure 2. Common features of microtubule-like structures (MTLs) and VIPP1 rings/rods. (A) Electron microscopy image of a microtubule-like structure (arrow) within a young spinach chloroplast. From Lawrence and Possingham (1984).Citation15 (B) Electron microscopy image after negative staining of recombinant Chlamydomonas VIPP1. The arrowhead indicates a VIPP1 ring. The scale is the same as that in .
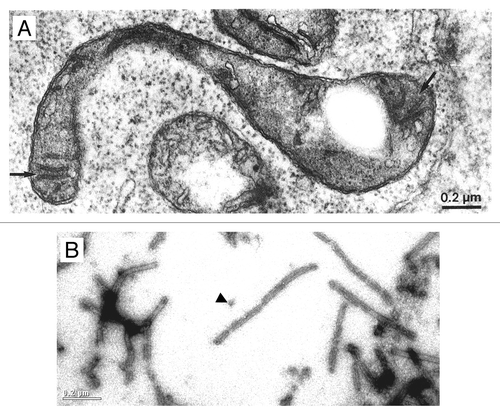
VIPP1 is conserved from cyanobacteria to higher plant chloroplasts. However, it is absent in some Prochlorococcus marinus strains that do contain thylakoid membranes. VIPP1 was found to form large ring-like structures of up to 2 MDa that have diameters of 25- to 37-nm and 12- to 17-fold symmetries.Citation19-Citation21 VIPP1 rings can further organize into rods, which may have lengths of up to 1.4 µmCitation21 (). Hence, VIPP1 rings/rods strongly resemble MTLs and the structures found within the cyanobacterial thylakoid centers. We hypothesize that MTLs might correspond to plastidic thylakoid centers and that these have VIPP1 as a central structural component (). This idea is supported by the following observations:
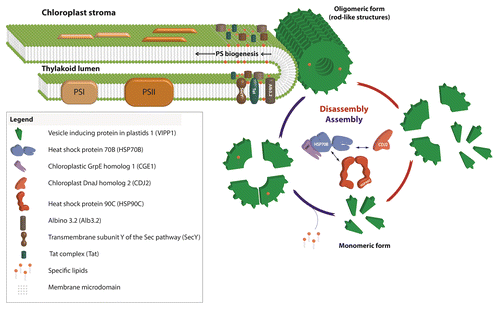
Figure 3. Model for the role of VIPP1 as structural but dynamic component of chloroplast thylakoid centers.
Already in the first publication on VIPP1 (then termed M30) the protein, VIPP1 by immunogold analysis was detected in clusters at thylakoids and at the envelope of pea chloroplasts.Citation22 GFP-tagged VIPP1 in Arabidopsis was found to organize into rod-shaped structures forming along the inner envelope.Citation8 Chloroplasts in Arabidopsis vipp1 knock-down lines had empty stromal spaces, as if the connection between the thylakoid network and the envelope was lost. Using immunofluorescence, VIPP1 within the chloroplast of Chlamydomonas cells was found to localize to distinct spots sometimes extending into rods.Citation6 In chloroplasts of VIPP1-amiRNA/RNAi cells, harboring only ~25% of wild-type VIPP levels, aberrant prolamellar body-like structures (PBLS) were consistently observed at the origin of multiple thylakoid membrane layers, which appear to coincide with the immunofluorescent VIPP1 spots. PBLSs were also found in dark-grown Chlamydomonas yellow-in-the-dark mutant cells that are deficient in the light-independent protochlorophyllide reductase.Citation23 These data suggested that a reduction in VIPP1 levels impairs thylakoid center formation and function, thereby leading to the aggregation of photosystem assembly intermediates in the PBLSs. Accordingly, levels of both photosystems were reduced by ~20% in Chlamydomonas VIPP1-RNAi strains, while thylakoid membranes in low light-grown cells appeared unaffected in their ultrastructure and lipid composition.Citation6 Similarly, depletion of VIPP1 in Synechocystis first led to a loss of the photosystems from thylakoid membranes before thylakoid membranes themselves were affected.Citation7 Further support for a role of VIPP1 within thylakoid centers in photosystem biogenesis comes from the observation that Chlamydomonas VIPP1 interacts with the membrane integrase ALB3.2, whose depletion by RNAi also led to a reduced accumulation of both photosystems.Citation24
VIPP1 rods and rings are dynamically (dis)assembled by the chloroplast HSP70B-CDJ2-CGE1 chaperone systemCitation21,Citation25 (). VIPP1 also interacts with chloroplast HSP90CCitation26 and co-suppression of the Arabidopsis HSP90C homolog resulted in a reduced accumulation of thylakoids and photosynthetic complexes and increased levels of VIPP1 oligomers.Citation27 These results indicate that the dynamic assembly and disassembly of VIPP1 by the chaperones is required for its function. Accordingly, GFP-tagged VIPP1 in osmotically stressed chloroplasts was found to be highly mobile in the stromal region, such that VIPP1 supercomplexes were formed at the expense of preexisting ones.Citation8 An interplay between VIPP1/HSP70B/CDJ2 and the membrane integrase ALB3.2 was also suggested by the finding that downregulation of ALB3.2 led to the upregulation of VIPP1 and the chaperones.Citation24
Finally, there is also evidence for a role of VIPP1 in supplying structural lipids, like phosphatidyl glycerol (PG), to PSIICitation6: 1) thylakoid membranes in Chlamydomonas VIPP1-RNAi cells grown on ammonium and exposed to high light intensities swell, suggesting a structural defect in these thylakoid membranes. This may be caused by a reduced ability of PSII to arrange in semicrystalline PSII-LHCII arrays caused by a reduced content in structural lipids in PSII. This scenario is also suggested by a reduced cooperativity of PSII centers in VIPP1-RNAi cells. 2) The integrity of the QA site of PSII is impaired in VIPP1-RNAi cells; in cyanobacterial PSII, PG depletion affected the integrity of the QB site.Citation28 3) PSII in VIPP1-RNAi cells is more thermosensitive, supporting the notion of a structural defect in this photosystem.
We speculate that VIPP1 might directly transport lipids to thylakoid centers during photosystem biogenesis. Alternatively, in analogy to eisosomes in the plasma membrane, VIPP1 might create microdomains enriched in specific lipid species, like PG (). Eisosomes are furrow-like invaginations of the plasma membrane enriched in sterols that in yeast are formed by the Pil1 and Lsp1 proteins.Citation29 Both proteins also form large rods resembling VIPP1 rods that are able to bind and tubulate liposomes in vitro.Citation30 The specific lipid environment created in microdomains formed by eisosomes attracts specific integral membrane proteins, e.g., the arginine transporter Can1, whose efficient function depends on the specific lipid environment present in these microdomains.Citation31 The enrichment of specific lipids like PG in a membrane microdomain putatively formed by VIPP1 may facilitate their incorporation as structural lipids during photosystem biogenesis. Moreover, translocases and integrases like ALB3, Sec, and TAT that are required for photosystem biogenesis may be enriched and their effective function dependent on the lipid environment in such microdomains (). In support for such a scenario, VIPP1 and its bacterial ancestor PspA improved bacterial TAT- and Sec-mediated protein secretion.Citation32,Citation33 The activity of the bacterial Sec translocase was shown to strictly require the presence of anionic phospholipids and is stimulated by non-bilayer lipids.Citation34 In this regard it is interesting that PspA was shown to bind PG and phosphatidylserineCitation35 and to interact with TatA.Citation36 Finally, the addition of recombinant VIPP1 to thylakoid membranes was shown to increase TAT-mediated protein import in vitro, although VIPP1 did not directly interact with import substrate or TAT components.Citation10
VIPP1 is strongly induced by high light, and the repair of PSII after photoinhibition was retarded in VIPP1-RNAi strains.Citation6 An attractive hypothesis is that VIPP1 newly synthesized at high light intensities would form additional thylakoid centers/microdomains to form sites for PSII repair. That plastidic HSP70B may catalyze this process is indicated by its strong inducibility by high lightCitation6 and the finding that HSP70B levels correlate with the ability of cells to recover from photoinhibition.Citation37
The working model presented here is speculative, but it will serve as a basis for targeted experiments to eventually elucidate the obviously very important function of VIPP1 in the biogenesis/repair of thylakoidal protein complexes.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We would like to thank Wiley-Blackwell for permission to reuse picture material originally published by ME Lawrence and JV Possingham in “Observations of microtubule-like structures within spinach plastids” in “Biology of the Cell”. This work was supported by the Deutsche Forschungsgemeinschaft (Schr 617/2–4 and 617/5–1).
References
- Eberhard S, Finazzi G, Wollman FA. The dynamics of photosynthesis. Annu Rev Genet 2008; 42:463 - 515; http://dx.doi.org/10.1146/annurev.genet.42.110807.091452; PMID: 18983262
- Adam Z, Charuvi D, Tsabari O, Knopf RR, Reich Z. Biogenesis of thylakoid networks in angiosperms: knowns and unknowns. Plant Mol Biol 2011; 76:221 - 34; http://dx.doi.org/10.1007/s11103-010-9693-5; PMID: 20859754
- Aseeva E, Ossenbühl F, Sippel C, Cho WK, Stein B, Eichacker LA, Meurer J, Wanner G, Westhoff P, Soll J, et al. Vipp1 is required for basic thylakoid membrane formation but not for the assembly of thylakoid protein complexes. Plant Physiol Biochem 2007; 45:119 - 28; http://dx.doi.org/10.1016/j.plaphy.2007.01.005; PMID: 17346982
- Kroll D, Meierhoff K, Bechtold N, Kinoshita M, Westphal S, Vothknecht UC, Soll J, Westhoff P. VIPP1, a nuclear gene of Arabidopsis thaliana essential for thylakoid membrane formation. Proc Natl Acad Sci U S A 2001; 98:4238 - 42; http://dx.doi.org/10.1073/pnas.061500998; PMID: 11274447
- Westphal S, Heins L, Soll J, Vothknecht UC. Vipp1 deletion mutant of Synechocystis: a connection between bacterial phage shock and thylakoid biogenesis?. Proc Natl Acad Sci U S A 2001; 98:4243 - 8; http://dx.doi.org/10.1073/pnas.061501198; PMID: 11274448
- Nordhues A, Schöttler MA, Unger AK, Geimer S, Schönfelder S, Schmollinger S, Rütgers M, Finazzi G, Soppa B, Sommer F, et al. Evidence for a role of VIPP1 in the structural organization of the photosynthetic apparatus in Chlamydomonas.. Plant Cell 2012; 24:637 - 59; http://dx.doi.org/10.1105/tpc.111.092692; PMID: 22307852
- Gao H, Xu X. Depletion of Vipp1 in Synechocystis sp. PCC 6803 affects photosynthetic activity before the loss of thylakoid membranes. FEMS Microbiol Lett 2009; 292:63 - 70; http://dx.doi.org/10.1111/j.1574-6968.2008.01470.x; PMID: 19222583
- Zhang L, Kato Y, Otters S, Vothknecht UC, Sakamoto W. Essential role of VIPP1 in chloroplast envelope maintenance in Arabidopsis.. Plant Cell 2012; 24:3695 - 707; http://dx.doi.org/10.1105/tpc.112.103606; PMID: 23001039
- Fuhrmann E, Gathmann S, Rupprecht E, Golecki J, Schneider D. Thylakoid membrane reduction affects the photosystem stoichiometry in the cyanobacterium Synechocystis sp. PCC 6803. Plant Physiol 2009; 149:735 - 44; http://dx.doi.org/10.1104/pp.108.132373; PMID: 19109411
- Lo SM, Theg SM. Role of vesicle-inducing protein in plastids 1 in cpTat transport at the thylakoid. Plant J 2012; 71:656 - 68; http://dx.doi.org/10.1111/j.1365-313X.2012.05020.x; PMID: 22487220
- Nickelsen J, Rengstl B, Photosystem II. Photosystem II assembly: from cyanobacteria to plants. Annu Rev Plant Biol 2013; 64:609 - 35; http://dx.doi.org/10.1146/annurev-arplant-050312-120124; PMID: 23451783
- Stengel A, Gügel IL, Hilger D, Rengstl B, Jung H, Nickelsen J. Initial steps of photosystem II de novo assembly and preloading with manganese take place in biogenesis centers in Synechocystis.. Plant Cell 2012; 24:660 - 75; http://dx.doi.org/10.1105/tpc.111.093914; PMID: 22319052
- Kunkel DD. Thylakoid centers: structures associated with the cyanobacterial photosynthetic membrane system. Arch Microbiol 1982; 133:97 - 9; http://dx.doi.org/10.1007/BF00413518
- van de Meene AM, Hohmann-Marriott MF, Vermaas WF, Roberson RW. The three-dimensional structure of the cyanobacterium Synechocystis sp. PCC 6803. Arch Microbiol 2006; 184:259 - 70; http://dx.doi.org/10.1007/s00203-005-0027-y; PMID: 16320037
- Lawrence ME, Possingham JV. Observations of microtubule-like structures within spinach plastids. Biol Cell 1984; 52:77 - 82
- Schnepf E. Plastidenstrukturen bei Passiflora.. Protoplasma 1961; 54:310 - 3; http://dx.doi.org/10.1007/BF01260360
- Pickett-Heaps JD. Microtubule-like structures in the growing plastids of two algae. Planta 1968; 81:193 - 200; http://dx.doi.org/10.1007/BF00417448
- Newcomb EH. Fine structure of protein-storing plastids in bean root tips. J Cell Biol 1967; 33:143 - 63; http://dx.doi.org/10.1083/jcb.33.1.143; PMID: 6033932
- Aseeva E, Ossenbühl F, Eichacker LA, Wanner G, Soll J, Vothknecht UC. Complex formation of Vipp1 depends on its α-helical PspA-like domain. J Biol Chem 2004; 279:35535 - 41; http://dx.doi.org/10.1074/jbc.M401750200; PMID: 15210715
- Fuhrmann E, Bultema JB, Kahmann U, Rupprecht E, Boekema EJ, Schneider D. The vesicle-inducing protein 1 from Synechocystis sp. PCC 6803 organizes into diverse higher-ordered ring structures. Mol Biol Cell 2009; 20:4620 - 8; http://dx.doi.org/10.1091/mbc.E09-04-0319; PMID: 19776353
- Liu C, Willmund F, Golecki JR, Cacace S, Hess B, Markert C, Schroda M. The chloroplast HSP70B-CDJ2-CGE1 chaperones catalyse assembly and disassembly of VIPP1 oligomers in Chlamydomonas.. Plant J 2007; 50:265 - 77; http://dx.doi.org/10.1111/j.1365-313X.2007.03047.x; PMID: 17355436
- Li HM, Kaneko Y, Keegstra K. Molecular cloning of a chloroplastic protein associated with both the envelope and thylakoid membranes. Plant Mol Biol 1994; 25:619 - 32; http://dx.doi.org/10.1007/BF00029601; PMID: 8061316
- Friedberg I, Goldberg I, Ohad I. A prolamellar body-like structure in Chlamydomonas reinhardi.. J Cell Biol 1971; 50:268 - 75; http://dx.doi.org/10.1083/jcb.50.1.268; PMID: 5563449
- Göhre V, Ossenbühl F, Crèvecoeur M, Eichacker LA, Rochaix JD. One of two alb3 proteins is essential for the assembly of the photosystems and for cell survival in Chlamydomonas.. Plant Cell 2006; 18:1454 - 66; http://dx.doi.org/10.1105/tpc.105.038695; PMID: 16679460
- Liu C, Willmund F, Whitelegge JP, Hawat S, Knapp B, Lodha M, Schroda M. J-domain protein CDJ2 and HSP70B are a plastidic chaperone pair that interacts with vesicle-inducing protein in plastids 1. Mol Biol Cell 2005; 16:1165 - 77; http://dx.doi.org/10.1091/mbc.E04-08-0736; PMID: 15635096
- Heide H, Nordhues A, Drepper F, Nick S, Schulz-Raffelt M, Haehnel W, Schroda M. Application of quantitative immunoprecipitation combined with knockdown and cross-linking to Chlamydomonas reveals the presence of vesicle-inducing protein in plastids 1 in a common complex with chloroplast HSP90C. Proteomics 2009; 9:3079 - 89; http://dx.doi.org/10.1002/pmic.200800872; PMID: 19526558
- Feng J, Fan P, Jiang P, Lv S, Chen X, Li Y. Chloroplast-targeted Hsp90 plays essential roles in plastid development and embryogenesis in Arabidopsis possibly linking with VIPP1. Physiol Plant 2013; http://dx.doi.org/10.1111/ppl.12083; PMID: 23875936
- Gombos Z, Várkonyi Z, Hagio M, Iwaki M, Kovács L, Masamoto K, Itoh S, Wada H. Phosphatidylglycerol requirement for the function of electron acceptor plastoquinone Q(B) in the photosystem II reaction center. Biochemistry 2002; 41:3796 - 802; http://dx.doi.org/10.1021/bi011884h; PMID: 11888298
- Strádalová V, Stahlschmidt W, Grossmann G, Blazíková M, Rachel R, Tanner W, Malinsky J. Furrow-like invaginations of the yeast plasma membrane correspond to membrane compartment of Can1. J Cell Sci 2009; 122:2887 - 94; http://dx.doi.org/10.1242/jcs.051227; PMID: 19638406
- Karotki L, Huiskonen JT, Stefan CJ, Ziółkowska NE, Roth R, Surma MA, Krogan NJ, Emr SD, Heuser J, Grünewald K, et al. Eisosome proteins assemble into a membrane scaffold. J Cell Biol 2011; 195:889 - 902; http://dx.doi.org/10.1083/jcb.201104040; PMID: 22123866
- Malinsky J, Opekarová M, Grossmann G, Tanner W. Membrane microdomains, rafts, and detergent-resistant membranes in plants and fungi. Annu Rev Plant Biol 2013; 64:501 - 29; http://dx.doi.org/10.1146/annurev-arplant-050312-120103; PMID: 23638827
- Vrancken K, De Keersmaeker S, Geukens N, Lammertyn E, Anné J, Van Mellaert L. pspA overexpression in Streptomyces lividans improves both Sec- and Tat-dependent protein secretion. Appl Microbiol Biotechnol 2007; 73:1150 - 7; http://dx.doi.org/10.1007/s00253-006-0571-7; PMID: 17106680
- DeLisa MP, Lee P, Palmer T, Georgiou G. Phage shock protein PspA of Escherichia coli relieves saturation of protein export via the Tat pathway. J Bacteriol 2004; 186:366 - 73; http://dx.doi.org/10.1128/JB.186.2.366-373.2004; PMID: 14702305
- van der Does C, Swaving J, van Klompenburg W, Driessen AJ. Non-bilayer lipids stimulate the activity of the reconstituted bacterial protein translocase. J Biol Chem 2000; 275:2472 - 8; http://dx.doi.org/10.1074/jbc.275.4.2472; PMID: 10644701
- Kobayashi R, Suzuki T, Yoshida M. Escherichia coli phage-shock protein A (PspA) binds to membrane phospholipids and repairs proton leakage of the damaged membranes. Mol Microbiol 2007; 66:100 - 9; http://dx.doi.org/10.1111/j.1365-2958.2007.05893.x; PMID: 17725563
- Mehner D, Osadnik H, Lünsdorf H, Brüser T. The Tat system for membrane translocation of folded proteins recruits the membrane-stabilizing Psp machinery in Escherichia coli.. J Biol Chem 2012; 287:27834 - 42; http://dx.doi.org/10.1074/jbc.M112.374983; PMID: 22689583
- Schroda M, Vallon O, Wollman FA, Beck CF. A chloroplast-targeted heat shock protein 70 (HSP70) contributes to the photoprotection and repair of photosystem II during and after photoinhibition. Plant Cell 1999; 11:1165 - 78; PMID: 10368186